Abstract
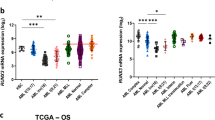
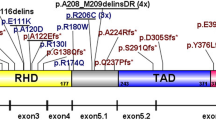
RUNX1A is the shortest and least expressed of the RUNX1 three main isoforms (A, B, C); despite this, the leukemogenic role of its overexpression has been clearly described. Several studies have shown RUNX1A involvement in different blood cancers and pilot observations in acute leukemia have been reported. In this context, we evaluated RUNX1 isoforms expression in a cohort of acute myeloid leukemia (AML) patients, finding overexpression of RUNX1A and RUNX1B, with higher median levels in thrombocytopenic cases. No difference was observed for RUNX1C. RUNX1A overexpression is higher in more immature AML phenotypes. According to the mutational profile, FLT3 internal tandem duplication (ITD) positive cases have the highest RUNX1A levels and the presence of FLT3-ITD was the only molecular variable able to influence RUNX1A expression. RUNX1A overexpression is disease-related, associated with a specific transcriptional profile, and reappears at relapse, with no clear kinetics except in FLT3-ITD cases. Overall, we demonstrate RUNX1A overexpression in AML and its association with the FLT3-ITD molecular subtype. Our data shed light on the dark side of RUNX1 deregulation, paving the way for further investigations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data generated in this study are available upon request from the corresponding author.
References
Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, et al. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–9.
Tanaka T, Tanaka K, Ogawa S, Kurokawa M, Mitani K, Nishida J, et al. An acute myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms. EMBO J. 1995;14:341–50.
Gialesaki S, Bräuer-Hartmann D, Issa H, Bhayadia R, Alejo-Valle O, Verboon L, et al. RUNX1 isoform disequilibrium promotes the development of trisomy 21–associated myeloid leukemia. Blood. 2023;141:1105–18.
Tsuzuki S, Seto M. Expansion of functionally defined mouse hematopoietic stem and progenitor cells by a short isoform of RUNX1/AML1. Blood. 2012;119:727–35.
Challen GA, Goodell MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol. 2010;38:403–16.
Liu X, Zhang Q, Zhang DE, Zhou C, Xing H, Tian Z, et al. Overexpression of an isoform of AML1 in acute leukemia and its potential role in leukemogenesis. Leukemia. 2009;23:739–45.
Sakurai H, Harada Y, Ogata Y, Kagiyama Y, Shingai N, Doki N, et al. Overexpression of RUNX1 short isoform has an important role in the development of myelodysplastic/myeloproliferative neoplasms. Blood Adv. 2017;1:1382–6.
Engvall M, Karlsson Y, Kuchinskaya E, Jörnegren Å, Mathot L, Pandzic T, et al. Familial platelet disorder due to germline exonic deletions in RUNX1: a diagnostic challenge with distinct alterations of the transcript isoform equilibrium. Leuk Lymphoma. 2022;63:2311–20.
Cabrerizo Granados D, Barbosa I, Baliakas P, Hellström-Lindberg E, Lundin V. The clinical phenotype of germline RUNX1 mutations in relation to the accompanying somatic variants and RUNX1 isoform expression. Genes Chromosom Cancer. 2023;62:672–7.
Cumbo C, Tarantini F, Anelli L, Zagaria A, Redavid I, Minervini CF, et al. IRF4 expression is low in Philadelphia negative myeloproliferative neoplasms and is associated with a worse prognosis. Exp Hematol Oncol. 2021;10:1–4.
Tarantini F, Cumbo C, Parciante E, Anelli L, Zagaria A, Coccaro N et al. IRF4 gene expression on the trail of molecular response: looking at chronic myeloid leukemia from another perspective. Acta Haematol. 2022. https://doi.org/10.1159/000527173.
Cumbo C, Tarantini F, Zagaria A, Anelli L, Minervini CF, Coccaro N, et al. Clonal hematopoiesis at the crossroads of inflammatory bowel diseases and hematological malignancies: a biological link?. Front Oncol. 2022;12:1–5.
Coccaro N, Zagaria A, Orsini P, Anelli L, Tota G, Casieri P, et al. RARA and RARG gene downregulation associated with EZH2 mutation in acute promyelocytic-like morphology leukemia. Hum Pathol. 2018;80:82–86.
Cumbo C, Orsini P, Tarantini F, Anelli L, Zagaria A, Tragni V, et al. TNFRSF13B gene mutation in familial acute myeloid leukemia: a new piece in the complex scenario of hereditary predisposition?. Hematol Oncol. 2023;41:942–6.
Murphy KM, Levis M, Hafez MJ, Geiger T, Cooper LC, Smith BD, et al. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5:96–102.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. 2017 https://doi.org/10.1182/blood-2016-08-733196.
Cumbo C, Tarantini F, Anelli L, Zagaria A, Specchia G, Musto P, et al. FLT3 mutational analysis in acute myeloid leukemia: advantages and pitfalls with different approaches. Blood Rev. 2022;54:100928.
Polak TB, van Rosmalen J, Dirven S, Herzig JK, Cloos J, Meshinchi S, et al. Association of FLT3-internal tandem duplication length with overall survival in acute myeloid leukemia: a systematic review and meta-analysis. Haematologica. 2022;107:2506–10.
Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–W221.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–30.
Lu J, Guo Y, Yin J, Chen J, Wang Y, Wang GG, et al. Structure-guided functional suppression of AML-associated DNMT3A hotspot mutations. Nat Commun. 2024;15:1–16.
Fu L, Fu H, Tian L, Xu K, Hu K, Wang J, et al. High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia. Oncotarget. 2016;7:15828–39.
Wang C, Tu Z, Cai X, Wang W, Davis AK, Nattamai K, et al. A critical role of RUNX1 in governing megakaryocyte-primed hematopoietic stem cell differentiation. Blood Adv. 2023;7:2590–605.
Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19.
Bennett JM, Catovsky D, Daniel M-T, Flandrin G, Galton DAG, Gralnick HR, et al. Proposals for the Classification of the Acute Leukaemias French-American-British (FAB) Co-operative Group. Br J Haematol. 1976;33:451–8.
Tsuzuki S, Hong D, Gupta R, Matsuo K, Seto M, Enver T. Isoform-specific potentiation of stem and progenitor cell engraftment by AML1/RUNX1. PLoS Med. 2007;4:0880–96.
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
Kellaway SG, Coleman DJL, Cockerill PN, Raghavan M, Bonifer C. Molecular basis of hematological disease caused by inherited or acquired RUNX1 mutations. Exp Hematol. 2022;111:1–12.
Davis AG, Einstein JM, Zheng D, Jayne ND, Fu XD, Tian B, et al. A CRISPR RNA-binding protein screen reveals regulators of RUNX1 isoform generation. Blood Adv. 2021;5:1310–23.
Komeno Y, Yan M, Matsuura S, Lam K, Lo MC, Huang YJ, et al. Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood. 2014;123:3760–9.
Fortier JM, Payton JE, Cahan P, Ley TJ, Walter MJ, Graubert TA. POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature. Leukemia. 2010;24:950–7.
Cheng CK, Li L, Cheng SH, Ng K, Chan NPH, Ip RKL, et al. Secreted-frizzled related protein 1 is a transcriptional repression target of the t(8;21) fusion protein in acute myeloid leukemia. Blood. 2011;118:6638–48.
De Bruijn M, Dzierzak E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017;129:2061–9.
Bellissimo DC, Speck NA. RUNX1 mutations in inherited and sporadic leukemia. Front Cell Dev Biol. 2017;5:1–11.
Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–82.
Cumbo C, Tota G, De Grassi A, Anelli L, Zagaria A, Coccaro N, et al. RUNX1 gene alterations characterized by allelic preference in adult acute myeloid leukemia. Leuk Lymphoma. 2021;0:1–9.
Zagaria A, Anelli L, Coccaro N, Tota G, Casieri P, Cellamare A, et al. 5′RUNX1-3′USP42 chimeric gene in acute myeloid leukemia can occur through an insertion mechanism rather than translocation and may be mediated by genomic segmental duplications. Mol Cytogenet. 2014;7:1–8.
Levanon D, Glusman G, Bangsow T, Ben-Asher E, Male DA, Avidan N, et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33.
Ghozi MC, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93:1935–40.
Cauchy P, James SR, Zacarias-Cabeza J, Ptasinska A, Imperato MR, Assi SA, et al. Chronic FLT3-ITD signaling in acute myeloid leukemia is connected to a specific chromatin signature. Cell Rep. 2015;12:821–36.
Behrens K, Maul K, Tekin N, Kriebitzsch N, Indenbirken D, Prassolov V, et al. RUNX1 cooperates with FLT3-ITD to induce leukemia. J Exp Med. 2017;214:737–52.
Acknowledgements
This work was supported by “Associazione Italiana contro le Leucemie (AIL)-BARI”.
Funding
This project received support from a grant awarded by Fondazione GIMEMA (Fondo per le Idee, 2021 edition).
Author information
Authors and Affiliations
Contributions
Conception and design of the study: CC and FA. Acquisition of data and/or analysis and interpretation of data: CC, FT, EP, LA, AZ, NC, GT, IR, MRC, AM, CFM, GS, PM, and FA. Clinical data acquisition: FT, VPG, and MD. Protein quantification: GB, AN, and FG. RNA-sequencing: MFC, FM, CT, SNC, BB and AT. Methylation analysis: PO and MG. Drafting of the manuscript: FA. All authors revised the manuscript for important intellectual content and approved the final version submitted for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The local Ethics Committee of “Azienda Ospedaliero Universitaria Policlinico di Bari” approved the study. Informed consent was obtained from all patients before study inclusion, in accordance with the Declaration of Helsinki. Patients’ records/information were anonymized and de-identified before analysis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cumbo, C., Tarantini, F., Parciante, E. et al. RUNX1A isoform is overexpressed in acute myeloid leukemia and is associated with FLT3 internal tandem duplications. Cancer Gene Ther 32, 963–972 (2025). https://doi.org/10.1038/s41417-025-00939-z
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41417-025-00939-z