Abstract
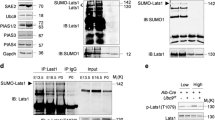
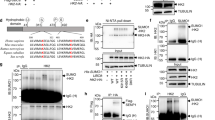
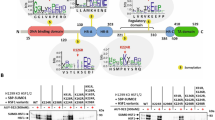
Kaiso is a member of the BTB/POZ zinc finger family, which is involved in cancer progression, cell cycle control, apoptosis, and WNT signaling. Depending on promoter context, it may function as either a transcriptional repressor or activator. Previous studies found that Kaiso might be SUMOylated due to heat shock, but the biological significance of Kaiso SUMOylation is unclear. Here, we find that K42 is the only amino acid within Kaiso that is modified with SUMO. Kaiso is monoSUMOylated at lysine 42 in cell lines of kidney origin under normal physiological conditions. SUMOylated Kaiso can activate transcription from exogenous methylated promoters, wherein the deSUMOylated form of the protein kept the ability to be a repressor. Rapid Kaiso deSUMOylation occurs in response to hyperosmotic stress and is reversible upon return to an isotonic environment. DeSUMOylation occurs within minutes in HEK293 cells treated with 100 mm NaCl and relaxes in 3 h even in a salt-containing medium. Genomic editing of Kaiso by conversion of K42 into R42 (K42R) in HEK293 cells that resulted in fully deSUMOylated endogenous protein led to misregulation of genes associated with ion transport, blood pressure, and the immune response. TRIM25 was significantly repressed in two K42R HEK293 clones. By a series of rescue experiments with K42R and KO HEK293 cells, we show that TRIM25 is a direct transcriptional target for Kaiso. In the absence of Kaiso, the level of TRIM25 is insensitive to hyperosmotic stress. Extending our observations to animal models, we show that in response to a high salt diet, Kaiso knockout mice are characterized by significantly higher blood pressure increases when compared to wild-type animals. Thus, we propose a novel biological role for Kaiso in the regulation of homeostasis.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C. et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–79.
Geiss-Friedlander R, Melchior F. Concepts in SUMOylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56.
Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–8.
Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH. et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteom. 2008;7:132–44.
Wei W, Yang P, Pang J, Zhang S, Wang Y, Wang MH. et al. A stress-dependent SUMO4 SUMOylation of its substrate proteins. Biochem Biophys Res Commun. 2008;375:454–9.
Liang YC, Lee CC, Yao YL, Lai CC, Schmitz ML, Yang WM. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci Rep. 2016;6:26509.
Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms?. Genes Dev. 2004;18:2046–59.
Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol. 2006;26:169–81.
Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–34.
Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS. et al. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Mol Cell. 2012;46:335–50.
Daniel JM, Spring CM, Crawford HC, Reynolds AB, Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–9.
Zhigalova NA, Sokolov AS, Prokhortchouk EB, Zhenilo SV. [S100A3 is a new target gene of Kaiso in mouse skin]. Mol Biol. 2015;49:362–5.
Zhenilo SV, Musharova OS, Pokhorchuk EB. [Transcription factor Kaiso does not interact with hydroxymethylated DNA within CTGCNA sequence context]. Mol Biol. 2013;47:522–5.
Pierre CC, Longo J, Bassey-Archibong BI, Hallett RM, Milosavljevic S, Beatty L. et al. Methylation-dependent regulation of hypoxia inducible factor-1 alpha gene expression by the transcription factor Kaiso. Biochim Biophys Acta. 2015;1849:1432–41.
Koh DI, Han D, Ryu H, Choi WI, Jeon BN, Kim MK. et al. KAISO, a critical regulator of p53-mediated transcription of CDKN1A and apoptotic genes. Proc Natl Acad Sci USA. 2014;111:15078–83.
Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–8.
Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A. et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24
Martin Caballero I, Hansen J, Leaford D, Pollard S, Hendrich BD. The methyl-CpG binding proteins Mecp2, Mbd2 and Kaiso are dispensable for mouse embryogenesis, but play a redundant function in neural differentiation. PLoS ONE. 2009;4:e4315
Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G. et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–8.
Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Prokhortchouk A, Sansom O, Selfridge J, Caballero IM, Salozhin S, Aithozhina D. et al. Kaiso-deficient mice show resistance to intestinal cancer. Mol Cell Biol. 2006;26:199–208.
Feng M, Deerhake ME, Keating R, Thaisz J, Xu L, Tsaih SW. et al. Genetic analysis of blood pressure in 8 mouse intercross populations. Hypertension. 2009;54:802–9.
Shumskaya VS, Zhigalova NA, Prokhorchouk AV, Prokhorchouk EB. Distribution of Kaiso protein in mouse tissues. Histochem Cell Biol. 2014;143:29–43.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Hilgarth RS, Sarge KD. Detection of SUMOylated proteins. Methods Mol Biol. 2005;301:329–38.
Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–23.
Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–9.
Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M. et al. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat Methods. 2007;4:245–50.
Ruzov A, Dunican DS, Prokhortchouk A, Pennings S, Stancheva I, Prokhortchouk E. et al. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development. 2004;131:6185–94.
Ruzov A, Savitskaya E, Hackett JA, Reddington JP, Prokhortchouk A, Madej MJ. et al. The non-methylated DNA-binding function of Kaiso is not required in early Xenopus laevis development. Development. 2009;136:729–38.
Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–23.
Capasso JM, Rivard CJ, Enomoto LM, Berl T. Chloride, not sodium, stimulates expression of the gamma subunit of Na/K-ATPase and activates JNK in response to hypertonicity in mouse IMCD3 cells. Proc Natl Acad Sci USA. 2003;100:6428–33.
Inoue S, Orimo A, Hosoi T, Kondo S, Toyoshima H, Kondo T. et al. Genomic binding-site cloning reveals an estrogen-responsive gene that encodes a RING finger protein. Proc Natl Acad Sci USA. 1993;90:11117–21.
Ikeda K, Orimo A, Higashi Y, Muramatsu M, Inoue S. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett. 2000;472:9–13.
Thomson SD, Ali S, Pickles L, Taylor J, Pace PE, Lymboura M. et al. Analysis of estrogen-responsive finger protein expression in benign and malignant human breast. Int J Cancer. 2001;91:152–8.
Qin Y, Cui H, Zhang H. Overexpression of TRIM25 in lung cancer regulates tumor cell progression. Technol Cancer Res Treat. 2016;15:707–15.
Zhang P, Elabd S, Hammer S, Solozobova V, Yan H, Bartel F, et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene. 2015;34(46):5729–38.
Zhigalova NA, Zhenilo SV, Aitkhozhina DS, Prokhorchuk EB. [Bifunctional role of domain zinc fingers of methyl-DNA-binding protein Kaiso]. Mol Biol. 2010;44(2):263–74.
Aillet F, Lopitz-Otsoa F, Egana I, Hjerpe R, Fraser P, Hay RT, et al. Heterologous SUMO-2/3-ubiquitin chains optimize IkappaBalpha degradation and NF-kappaB activity. PLoS ONE. 2012;7(12):e51672.
Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2(2):233–9.
Blattler A, Yao L, Wang Y, Ye Z, Jin VX, Farnham PJ. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenet Chromat. 2013;6(1):13.
Brocker C, Thompson DC, Vasiliou V. The role of hyperosmotic stress in inflammation and disease. Biomol Concepts. 2012;3(4):345–64.
Acknowledgements
We thank Dr. B. Hendrich for providing MBD2−/−, Kaiso−/y fibroblasts cell line, Dr. R. Niedenthal for SUMO1-GFP, pcDNA3-UBC9 plasmids, Dr. P. Sergiev for HA-agarose. We thank Dr. A. Reynolds for providing with anti-Kaiso polyclonal rabbit. We thank Dr. I. Petrushanko and Dr. S. Chumakov for assistance in flow cytometry. We thank Dr. S. Salozhin for fruitful discussion of the work.
Funding
The work was supported by RNF 14-14-01202 and part of the work related with mice was funded by budget from project No. 0324-2018-0016 of the Federal Research Center “Institute of Cytology and Genetics” SB RAS.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by V. D'Angiolella.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhenilo, S., Deyev, I., Litvinova, E. et al. DeSUMOylation switches Kaiso from activator to repressor upon hyperosmotic stress. Cell Death Differ 25, 1938–1951 (2018). https://doi.org/10.1038/s41418-018-0078-7
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-018-0078-7
This article is cited by
-
SUMOylation of TRIM28 is positively modulated by the BTB/POZ domain of Kaiso
Molecular Biology Reports (2025)
-
Dissecting the Kaiso binding profile in clear renal cancer cells
Epigenetics & Chromatin (2024)
-
Expressional variations of Kaiso: an association with pathological characteristics and field cancerization of OSCC
BMC Cancer (2022)
-
Identifying molecular targets for reverse aging using integrated network analysis of transcriptomic and epigenomic changes during aging
Scientific Reports (2021)