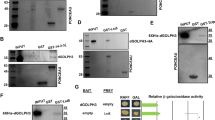
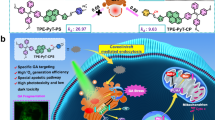
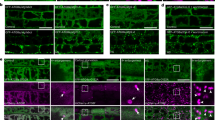
Abstract
LC3 is a protein that can associate with autophagosomes, autolysosomes, and phagosomes. Here, we show that LC3 can also redistribute toward the damaged Golgi apparatus where it clusters with SQSTM1/p62 and lysosomes. This organelle-specific relocation, which did not involve the generation of double-membraned autophagosomes, could be observed after Golgi damage was induced by various strategies, namely (i) laser-induced localized cellular damage, (ii) local expression of peroxidase and exposure to peroxide and diaminobenzidine, (iii) treatment with the Golgi-tropic photosensitizer redaporfin and light, (iv) or exposure to the Golgi-tropic anticancer peptidomimetic LTX-401. Mechanistic exploration led to the conclusion that both reactive oxygen species-dependent and -independent Golgi damage induces a similar phenotype that depended on ATG5 yet did not depend on phosphatidylinositol-3-kinase catalytic subunit type 3 and Beclin-1. Interestingly, knockout of ATG5 sensitized cells to Golgi damage-induced cell death, suggesting that the pathway culminating in the relocation of LC3 to the damaged Golgi may have a cytoprotective function.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222.
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8.
Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94.
Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100.
Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18.
Birgisdottir AB, Lamark T, Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013;126(Pt 15):3237–47.
Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–57.
Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Cell death assays for drug discovery. Nat Rev Drug Discov. 2011;10:221–37.
BenYounes A, Tajeddine N, Tailler M, Malik SA, Shen S, Metivier D, et al. A fluorescence-microscopic and cytofluorometric system for monitoring the turnover of the autophagic substrate p62/SQSTM1. Autophagy. 2011;7:883–91.
Martinez J, Malireddi RK, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17:893–906.
Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–79.
Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, et al. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9:493–8.
Jollivet F, Raposo G, Dimitrov A, Sougrat R, Goud B, Perez F. Analysis of de novo Golgi complex formation after enzyme-based inactivation. Mol Biol Cell. 2007;18:4637–47.
Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435.
Tasdemir E, Galluzzi L, Maiuri MC, Criollo A, Vitale I, Hangen E, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol Biol. 2008;445:29–76.
Gomes-da-Silva LC, Zhao L, Bezu L, Zhou H, Sauvat A, Liu P, et al. Photodynamic therapy with redaporfin targets the endoplasmic reticulum and Golgi apparatus. EMBO J. 2018; in press.
Garg AD, Krysko DV, Verfaillie T, Kaczmarek A, Ferreira GB, Marysael T, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 2012;31:1062–79.
Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–4.
Saenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, et al. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol. 2009;5:157–65.
Zhou H, Sauvat A, Gomes-da-Silva LC, Durand S, Forveille S, Iribarren K, et al. The oncolytic compound LTX-401 targets the Golgi apparatus. Cell Death Differ. 2016;23:2031–41.
Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–70.
Niso-Santano M, Malik SA, Pietrocola F, Bravo-San Pedro JM, Marino G, Cianfanelli V, et al. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015;34:1025–41.
Shen S, Kepp O, Kroemer G. The end of autophagic cell death? Autophagy. 2012;8:1–3.
Wei Y, Chiang WC, Sumpter R Jr, Mishra P, Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–38. e210
Pickles S, Vigie P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–R185.
Rubio N, Coupienne I, Di Valentin E, Heirman I, Grooten J, Piette J, et al. Spatiotemporal autophagic degradation of oxidatively damaged organelles after photodynamic stress is amplified by mitochondrial reactive oxygen species. Autophagy. 2012;8:1312–24.
Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–69.
Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58.
Slot JW, Geuze HJ, Gigengack S, Lienhard GE, James DE. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J Cell Biol. 1991;113:123–35.
Acknowledgements
The plasmid pEGFP-LC3 wt was a gift from Dr Noboru Mizushima (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). The pGalT-EGFP plasmid was a kind gift from J. White, EMBL, Heidelberg, Germany. The pDest-mCherry-p62 plasmid was a kind gift from Terje Johansen. The pExp-Dh1-GFP-LC3-Ef1-Puro for lentiviral production was a kind gift of François-Xavier Gobert and Philippe Benaroch. Luzitin SA provided redaporfin and F2BOH. Sabrina Forveille for her help on assessing colonies numbers. LCGdS is supported by Portuguese Science Foundation (ref. SFRH/BPD/93562/2013). GK is supported by the Ligue contre le Cancer (équipe labelisée); Agence National de la Recherche (ANR)—Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Institut National du Cancer (INCa); Institut Universitaire de France; Fondation pour la Recherche Médicale (FDM20140630126 and FDM 40739); the European Commission (ArtForce); the European Research Council (ERC); the LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumière, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI). FP is supported by the Centre National de la Recherche Scientifique (CNRS), the Institut Curie and the LabEx CelTisPhyBio (ANR-10-LBX-0038 part of the IDEX PSL no ANR-10-IDEX-0001–02).
Author information
Authors and Affiliations
Contributions
L.C.G.d.S. performed the experiments with redaporfin-PDT and LTX-401 and A.J.J. performed the experiments of Golgi damage using laser photodamage and HRP-ManII-expressing cells. A.S. developed R scripts used for analysis. S.S., S.D., and M.S. performed electron microscopy, Ø.R. and B.S. provided LTX-401. L.A., O.K., G.K., and F.P. designed the study. G.K. and F.P. wrote the paper.
Corresponding authors
Ethics declarations
Disclosure
BS and ØR are full-time employees of Lytix Biopharma and this study was partially funded by Lytix Biopharma.
Additional information
Edited by H.-U. Simon
Rights and permissions
About this article
Cite this article
Gomes-da-Silva, L.C., Jimenez, A.J., Sauvat, A. et al. Recruitment of LC3 to damaged Golgi apparatus. Cell Death Differ 26, 1467–1484 (2019). https://doi.org/10.1038/s41418-018-0221-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-018-0221-5
This article is cited by
-
Non-autophagic Golgi-LC3 lipidation facilitates TFE3 stress response against Golgi dysfunction
The EMBO Journal (2024)
-
A non-canonical role of ATG8 in Golgi recovery from heat stress in plants
Nature Plants (2023)
-
ATG8 keeps Golgi in shape after the heat
Nature Plants (2023)
-
ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy
Nature Structural & Molecular Biology (2022)
-
Screening and identification of potential biomarkers and therapeutic drugs in melanoma via integrated bioinformatics analysis
Investigational New Drugs (2021)