Abstract
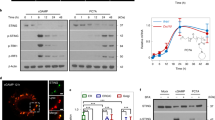
STING (stimulator of interferon genes) is a central molecule that binds to cyclic dinucleotides produced by the cyclic GMP-AMP synthase (cGAS) to activate innate immunity against microbial infection. Here we report that STING harbors classic LC-3 interacting regions (LIRs) and mediates autophagy through its direct interaction with LC3. We observed that poly(dA:dT), cGAMP, and HSV-1 induced STING-dependent autophagy and degradation of STING immediately after TBK1 activation. STING induces non-canonical autophagy that is dependent on ATG5, whereas other autophagy regulators such as Beclin1, Atg9a, ULK1, and p62 are dispensable. LIR mutants of STING abolished its interaction with LC3 and its activation of autophagy. Also, mutants that abolish STING dimerization and cGAMP-binding diminished the STING-LC3 interaction and subsequent autophagy, suggesting that STING activation is indispensable for autophagy induction. Our results thus uncover dual functions of STING in activating the immune response and autophagy, and suggest that STING is involved in ensuring a measured innate immune response.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Sun LJ, Wu JX, Du FH, Chen X, Chen ZJJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91.
Wu JX, Sun LJ, Chen X, Du FH, Shi HP, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–30.
Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8.
Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–26.
Sun WX, Li Y, Chen L, Chen HH, You FP, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA. 2009;106:8653–8.
Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao FC, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–50.
Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–6.
Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92.
McWhirter SM, tenOever BR, Maniatis T. Connecting mitochondria and innate immunity. Cell. 2005;122:645–7.
Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–9.
Yin Q, Tian Y, Kabaleeswaran V, Jiang XM, Tu DQ, Eck MJ, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–45.
Liu SQ, Cai X, Wu JX, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630.
Tanaka Y, Chen ZJJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20.
Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–76.
Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–55.
Zhong B, Zhang L, Lei CQ, Li Y, Mao AP, Yang Y, et al. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407.
Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014;10:e1004358.
Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–9.
Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243–51.
Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–90.
Jiao M, Ren F, Zhou L, Zhang X, Zhang L, Wen T, et al. Peroxisome proliferator-activated receptor alpha activation attenuates the inflammatory response to protect the liver from acute failure by promoting the autophagy pathway. Cell Death Dis. 2014;5:e1397.
Ren F, Zhang L, Zhang X, Shi H, Wen T, Bai L, et al. Inhibition of glycogen synthase kinase 3beta promotes autophagy to protect mice from acute liver failure mediated by peroxisome proliferator-activated receptor alpha. Cell Death Dis. 2016;7:e2151.
Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–34.
Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–42.
Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–44.
Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33.
Bhatelia K, Singh K, Prajapati P, Sripada L, Roy M, Singh R. MITA modulated autophagy flux promotes cell death in breast cancer cells. Cell Signal. 2017;35:73–83.
Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171:809–23.
Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–98.
Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018; 37:e97858.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45.
Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–85.
Jacomin AC, Samavedam S, Promponas V, Nezis IP. iLIR database: a web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945–53.
Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–86.
Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, et al. Structure-function analysis of STING activation by c[G(2’,5’)pA(3’,5’)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–62.
Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–4.
Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017;27:184–201.
Young ARJ, Chan EYW, Hu XW, Koch R, Crawshaw SG, High S, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900.
Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–99.
Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85.
Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–15.
Lim J, Murthy A. Controlling inflammation by selective autophagy. Cell Death Differ. 2018;25:825–7.
Cheng J, Liao Y, Xiao L, Wu R, Zhao S, Chen H, et al. Autophagy regulates MAVS signaling activation in a phosphorylation-dependent manner in microglia. Cell Death Differ. 2017;24:276–87.
Fang R, Wang C, Jiang Q, Lv M, Gao P, Yu X, et al. NEMO-IKKbeta are essential for IRF3 and NF-kappaB activation in the cGAS-STING pathway. J Immunol. 2017;199:3222–33.
Acknowledgements
We are grateful to Professor Shuai Chen (Sun Yat-sen University Cancer Center, Guangzhou, China) for sharing the Flag-tagged STING, IFN-β luciferase reporter gene and pRL-TK plasmids. We thank Professor Hongyu Deng (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China) for providing HSV-1 virus. We thank Professor Pu Gao (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China) for providing the structural analysis of STING LIR motifs. We would be grateful to Professor Pu Gao and Jingpeng Zhu (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China) for technical help with protein purification. We thank Ms. Yinzi Ma and Mr. Pengyan Xia (Institute of Zoology, Chinese Academy of Sciences, Beijing, China) for their help with electron microscopy assays. We wish to thank Dr. Qing Zhong (University of Texas Southwestern Medical Center, Dallas, TX, USA) for his suggestions and critical reading of the manuscript. This work was supported by grants from the Special Fund for Strategic Pilot Technology Chinese Academy of Sciences (QYZDJ-SSW-SMC004), Fund for Strategic Pilot Technology Chinese Academy of Sciences (XDPB1002), the Natural Science Foundation of China (31790404), the Beijing Natural Science Foundation of China (5161002), 973 Program Projects (2015CB856303).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, D., Wu, H., Wang, C. et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ 26, 1735–1749 (2019). https://doi.org/10.1038/s41418-018-0251-z
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-018-0251-z
This article is cited by
-
Lactiplantibacillus plantarum extracellular vesicles exert anti-PEDV effects through STING-dependent autophagy
BMC Microbiology (2025)
-
The cGAS-STING pathway: a dual regulator of immune response in cancer and therapeutic implications
Journal of Translational Medicine (2025)
-
The neuroimmune nexus: unraveling the role of the mtDNA-cGAS-STING signal pathway in Alzheimer’s disease
Molecular Neurodegeneration (2025)
-
TRIM23 mediates cGAS-induced autophagy in anti-HSV defense
Nature Communications (2025)
-
Regulation of cGAS–STING signalling and its diversity of cellular outcomes
Nature Reviews Immunology (2025)