Abstract
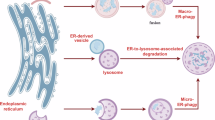
Autophagy regulates the degradation of unnecessary or dysfunctional cellular components. This catabolic process requires the formation of a double-membrane vesicle, the autophagosome, that engulfs the cytosolic material and delivers it to the lysosome. Substrate specificity is achieved by autophagy receptors, which are characterized by the presence of at least one LC3-interaction region (LIR) or GABARAP-interaction motif (GIM). Only recently, several receptors that mediate the specific degradation of endoplasmic reticulum (ER) components via autophagy have been identified (the process known as ER-phagy or reticulophagy). Here, we give an update on the current knowledge about the role of ER-phagy receptors in health and disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214.
Goyal U, Blackstone C. Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta. 2013;1833:2492–8.
Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, et al. Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science. 2016;354:aaf3928.
Beetz C, Koch N, Khundadze M, Zimmer G, Nietzsche S, Hertel N, et al. A spastic paraplegia mouse model reveals REEP1-dependent ER shaping. J Clin Investig. 2013;123:4273–82.
Hubner CA, Kurth I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain. 2014;137:3109–21.
Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–88.
Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–86.
Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6.
Fregno I, Molinari M. Endoplasmic reticulum turnover: ER-phagy and other flavors in selective and non-selective ER clearance. F1000 Res. 2018;7:454.
Fregno I, Molinari M. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit Rev Biochem Mol Biol. 2019;54:153–63.
Sun Z, Brodsky JL. Protein quality control in the secretory pathway. J Cell Biol. 2019;218:3171–87.
Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–41.
Henne WM, Liou J, Emr SD. Molecular mechanisms of inter-organelle ER-PM contact sites. Curr Opin Cell Biol. 2015;35:123–30.
Bolender RP, Weibel ER. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J Cell Biol. 1973;56:746–61.
Rogov VV, Stolz A, Ravichandran AC, Rios-Szwed DO, Suzuki H, Kniss A, et al. Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep. 2018;19:e47268.
Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501.
Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, et al. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II). Hum Mol Genet. 2007;16:618–29.
Dikic I. Open questions: why should we care about ER-phagy and ER remodelling? BMC Biol. 2018;16:131.
Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–8.
Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016;18:1173–84.
Grumati P, Morozzi G, Holper S, Mari M, Harwardt MI, Yan R, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017;6:e25555.
Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, et al. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell. 2018;44:217–32. e211.
Chen Q, Xiao Y, Chai P, Zheng P, Teng J, Chen J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr Biol. 2019;29:846–55. e846.
An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, Harper JW. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol Cell. 2019;74:891–908. e810.
Chino H, Hatta T, Natsume T, Mizushima N. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol Cell. 2019;74:909–21. e906.
Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–8.
Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–62.
Fregno I, Fasana E, Bergmann TJ, Raimondi A, Loi M, Solda T, et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018;37:e99259.
Forrester A, De Leonibus C, Grumati P, Fasana E, Piemontese M, Staiano L, et al. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 2019;38:e99847.
Cui Y, Parashar S, Zahoor M, Needham PG, Mari M, Zhu M, et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science. 2019;365:53–60.
Chiramel AI, Dougherty JD, Nair V, Robertson SJ, Best SM. FAM134B, the selective autophagy receptor for endoplasmic reticulum turnover, inhibits replication of ebola virus strains makona and mayinga. J Infect Dis. 2016;214(suppl 3):S319–25.
Moretti J, Roy S, Bozec D, Martinez J, Chapman JR, Ueberheide B, et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171:809–23. e813.
Liang JR, Lingeman E, Ahmed S, Corn JE. Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol. 2018;217:3354–67.
Bhaskara RM, Grumati P, Garcia-Pardo J, Kalayil S, Covarrubias-Pinto A, Chen W, et al. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun. 2019;10:2370.
Schultz ML, Krus KL, Kaushik S, Dang D, Chopra R, Qi L, et al. Coordinate regulation of mutant NPC1 degradation by selective ER autophagy and MARCH6-dependent ERAD. Nat Commun. 2018;9:3671.
Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41:1179–81.
Davidson G, Murphy S, Polke J, Laura M, Salih M, Muntoni F, et al. Frequency of mutations in the genes associated with hereditary sensory and autonomic neuropathy in a UK cohort. J Neurol. 2012;259:1673–85.
Ilgaz Aydinlar E, Rolfs A, Serteser M, Parman Y. Mutation in FAM134B causing hereditary sensory neuropathy with spasticity in a Turkish family. Muscle Nerve. 2014;49:774–5.
Murphy SM, Davidson GL, Brandner S, Houlden H, Reilly MM. Mutation in FAM134B causing severe hereditary sensory neuropathy. J Neurol Neurosurg Psychiatry. 2012;83:119–20.
Tang WK, Chui CH, Fatima S, Kok SH, Pak KC, Ou TM, et al. Oncogenic properties of a novel gene JK-1 located in chromosome 5p and its overexpression in human esophageal squamous cell carcinoma. Int J Mol Med. 2007;19:915–23.
Haque MH, Gopalan V, Chan KW, Shiddiky MJ, Smith RA, Lam AK. Identification of Novel FAM134B (JK1) Mutations in Oesophageal Squamous Cell Carcinoma. Sci Rep. 2016;6:29173.
Kasem K, Gopalan V, Salajegheh A, Lu CT, Smith RA, Lam AK. The roles of JK-1 (FAM134B) expressions in colorectal cancer. Exp Cell Res. 2014;326:166–73.
Islam F, Gopalan V, Lam AK. RETREG1 (FAM134B): a new player in human diseases: 15 years after the discovery in cancer. J Cell Physiol. 2018;233:4479–89.
Melchiotti R, Puan KJ, Andiappan AK, Poh TY, Starke M, Zhuang L, et al. Genetic analysis of an allergic rhinitis cohort reveals an intercellular epistasis between FAM134B and CD39. BMC Med Genet. 2014;15:73.
Kong M, Kim Y, Lee C. A strong synergistic epistasis between FAM134B and TNFRSF19 on the susceptibility to vascular dementia. Psychiatr Genet. 2011;21:37–41.
Lennemann NJ, Coyne CB. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy. 2017;13:322–32.
Shi Q, Ge Y, Sharoar MG, He W, Xiang R, Zhang Z, et al. Impact of RTN3 deficiency on expression of BACE1 and amyloid deposition. J Neurosci. 2014;34:13954–62.
Cunningham CN, Williams JM, Knupp J, Arunagiri A, Arvan P, Tsai B. Cells deploy a two-pronged strategy to rectify misfolded proinsulin aggregates. Mol Cell. 2019;75:442–56. e444.
Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–61.
Chen Q, Teng J, Chen J. ATL3, a cargo receptor for reticulophagy. Autophagy. 2019;15:1465–6.
Kornak U, Mademan I, Schinke M, Voigt M, Krawitz P, Hecht J, et al. Sensory neuropathy with bone destruction due to a mutation in the membrane-shaping atlastin GTPase 3. Brain. 2014;137:683–92.
Krols M, Asselbergh B, De Rycke R, De Winter V, Seyer A, Muller FJ, et al. Sensory neuropathy-causing mutations in ATL3 affect ER-mitochondria contact sites and impair axonal mitochondrial distribution. Hum Mol Genet. 2019;28:615–27.
Xu H, Zhang C, Cao L, Song J, Xu X, Zhang B, et al. ATL3 gene mutation in a Chinese family with hereditary sensory neuropathy type 1F. J Peripher Nerv Syst. 2019;24:150–5.
Behrendt L, Kurth I, Kaether C. A disease causing ATLASTIN 3 mutation affects multiple endoplasmic reticulum-related pathways. Cell Mol Life Sci. 2019;76:1433–45.
Niu L, Ma T, Yang F, Yan B, Tang X, Yin H, et al. Atlastin-mediated membrane tethering is critical for cargo mobility and exit from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2019;116:14029–38.
Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU, et al. Mammalian Sec61 is associated with Sec62 and Sec63. J Biol Chem. 2000;275:14550–7.
Linxweiler M, Schick B, Zimmermann R. Let’s talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine. Signal Transduct Target Ther. 2017;2:17002.
Greiner M, Kreutzer B, Jung V, Grobholz R, Hasenfus A, Stohr RF, et al. Silencing of the SEC62 gene inhibits migratory and invasive potential of various tumor cells. Int J Cancer. 2011;128:2284–95.
Greiner M, Kreutzer B, Lang S, Jung V, Cavalie A, Unteregger G, et al. Sec62 protein level is crucial for the ER stress tolerance of prostate cancer. Prostate. 2011;71:1074–83.
Linxweiler M, Bochen F, Schick B, Wemmert S, Al Kadah B, Greiner M, et al. Identification of SEC62 as a potential marker for 3q amplification and cellular migration in dysplastic cervical lesions. BMC Cancer. 2016;16:676.
Linxweiler M, Linxweiler J, Barth M, Benedix J, Jung V, Kim YJ, et al. Sec62 bridges the gap from 3q amplification to molecular cell biology in non-small cell lung cancer. Am J Pathol. 2012;180:473–83.
Wemmert S, Lindner Y, Linxweiler J, Wagenpfeil S, Bohle R, Niewald M, et al. Initial evidence for Sec62 as a prognostic marker in advanced head and neck squamous cell carcinoma. Oncol Lett. 2016;11:1661–70.
Bergmann TJ, Fumagalli F, Loi M, Molinari M. Role of SEC62 in ER maintenance: a link with ER stress tolerance in SEC62-overexpressing tumors? Mol Cell Oncol. 2017;4:e1264351.
Kostenko EV, Olabisi OO, Sahay S, Rodriguez PL, Whitehead IP. Ccpg1, a novel scaffold protein that regulates the activity of the Rho guanine nucleotide exchange factor Dbs. Mol Cell Biol. 2006;26:8964–75.
Schwartzlow C, Kazamel M. Hereditary sensory and autonomic neuropathies: adding more to the classification. Curr Neurol Neurosci Rep. 2019;19:52.
Eggermann K, Gess B, Hausler M, Weis J, Hahn A, Kurth I. Hereditary Neuropathies. Dtsch Arztebl Int. 2018;115:91–7.
Fink JK. Hereditary spastic paraplegia. Curr Neurol Neurosci Rep. 2006;6:65–76.
Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29:326–31.
Guelly C, Zhu PP, Leonardis L, Papic L, Zidar J, Schabhuttl M, et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am J Hum Genet. 2011;88:99–105.
Montenegro G, Rebelo AP, Connell J, Allison R, Babalini C, D’Aloia M, et al. Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Investig. 2012;122:538–44.
Zou Y, He W, Wang K, Han H, Xiao T, Chen X, et al. Identification of rare RTN3 variants in Alzheimer’s disease in Han Chinese. Hum Genet. 2018;137:141–50.
Heath JE, Siedlak SL, Zhu X, Lee HG, Thakur A, Yan R, et al. Widespread distribution of reticulon-3 in various neurodegenerative diseases. Neuropathology. 2010;30:574–9.
Kasem K, Gopalan V, Salajegheh A, Lu CT, Smith RA, Lam AK. JK1 (FAM134B) gene and colorectal cancer: a pilot study on the gene copy number alterations and correlations with clinicopathological parameters. Exp Mol Pathol. 2014;97:31–36.
Viswanath P, Radoul M, Izquierdo-Garcia JL, Luchman HA, Gregory Cairncross J, Pieper RO, et al. Mutant IDH1 gliomas downregulate phosphocholine and phosphoethanolamine synthesis in a 2-hydroxyglutarate-dependent manner. Cancer Metabol. 2018;6:3.
Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–33.
Acknowledgements
We are grateful to Paolo Grumati, Andrea Gubas, Ingo Kurth, and Antje-Kathrin Huebner for critical comments and insightful discussions. This work was partially supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project number 259130777—SFB 1177 and DFG-funded Research Unit FOR 2625 on Mechanisms of Lysosomal Homeostasis, the European Research Council (ERC) grant 742720 UbBAC, Cardio-Pulmonary Institute (CPI), EXC 2026, Project ID: 390649896, and the LOEWE program on Ubiquitin Networks (Ub-Net), State of Hesse, Germany to ID and by German Research Foundation HU 800/6–2, HU 800/13–1, the DFG-funded Research Unit FOR 2625 on Mechanisms of Lysosomal Homeostasis and the SP6 within the DFG-funded research training group 1715 to CAH. CAH and ID envisioned and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by F. Pentimalli
Rights and permissions
About this article
Cite this article
Hübner, C.A., Dikic, I. ER-phagy and human diseases. Cell Death Differ 27, 833–842 (2020). https://doi.org/10.1038/s41418-019-0444-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-019-0444-0
This article is cited by
-
The role of ER-associated degradation and ER-phagy in health and disease
Signal Transduction and Targeted Therapy (2026)
-
ER-phagy receptors: structural mechanisms in selective ER degradation and disease implications
Acta Pharmacologica Sinica (2026)
-
MARCH6 Confers Protection Against Endoplasmic Reticulum Autophagy in Gliomas by Destabilizing FAM134B
Neurochemical Research (2026)
-
The many connections of UFMylation with Alzheimer’s disease: a comprehensive review
Molecular Neurodegeneration (2025)
-
ER-phagy mediates the anti-tumoral synergism between HDAC inhibition and chemotherapy
Cell Communication and Signaling (2025)