Abstract
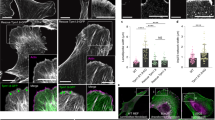
Autophagy ensures the turnover of cytoplasm and requires the coordinated action of Atg proteins, some of which also have moonlighting functions in higher eukaryotes. Here we show that the transmembrane protein Atg9 is required for female fertility, and its loss leads to defects in actin cytoskeleton organization in the ovary and enhances filopodia formation in neurons in Drosophila. Atg9 localizes to the plasma membrane anchor points of actin cables and is also important for the integrity of the cortical actin network. Of note, such phenotypes are not seen in other Atg mutants, suggesting that these are independent of autophagy defects. Mechanistically, we identify the known actin regulators profilin and Ena/VASP as novel binding partners of Atg9 based on microscopy, biochemical, and genetic interactions. Accordingly, the localization of both profilin and Ena depends on Atg9. Taken together, our data identify a new and unexpected role for Atg9 in actin cytoskeleton regulation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30.
Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93:1015–34.
Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40.
Orsi A, Razi M, Dooley H, Robinson D, Weston A, Collinson L, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, is required for autophagy. Mol Biol Cell. 2012;23:1860–73.
Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–99.
Barth JM, Hafen E, Kohler K. The lack of autophagy triggers precocious activation of Notch signaling during Drosophila oogenesis. BMC Dev Biol. 2012;12:35.
Barth JM, Szabad J, Hafen E, Kohler K. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 2011;18:915–24.
Varga K, Nagy P, Arsikin Csordas K, Kovacs AL, Hegedus K, Juhasz G. Loss of Atg16 delays the alcohol-induced sedation response via regulation of Corazonin neuropeptide production in Drosophila. Sci Rep. 2016;6:34641.
Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–6.
Wen JK, Wang YT, Chan CC, Hsieh CW, Liao HM, Hung CC, et al. Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. Elife. 2017;6:1–22.
McLean PF, Cooley L. Bridging the divide: illuminating the path of intercellular exchange through ring canals. Fly. 2014;8:13–8.
Revenu C, Athman R, Robine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol. 2004;5:635–46.
Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–28.
Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, et al. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–62.
Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA. 2006;103:7694–9.
Sechi AS, Wehland J. ENA/VASP proteins: multifunctional regulators of actin cytoskeleton dynamics. Front Biosci: a J virtual Libr. 2004;9:1294–310.
Takats S, Glatz G, Szenci G, Boda A, Horvath GV, Hegedus K, et al. Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet. 2018;14:e1007359.
Lorincz P, Lakatos Z, Varga A, Maruzs T, Simon-Vecsei Z, Darula Z, et al. MiniCORVET is a Vps8-containing early endosomal tether in Drosophila. Elife. 2016;5:e14226.
Hegedus K, Takats S, Boda A, Jipa A, Nagy P, Varga K, et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell. 2016;27:3132–42.
Nagy P, Hegedus K, Pircs K, Varga A, Juhasz G. Different effects of Atg2 and Atg18 mutations on Atg8a and Atg9 trafficking during starvation in Drosophila. FEBS Lett. 2014;588:408–13.
Kim M, Sandford E, Gatica D, Qiu Y, Liu X, Zheng Y, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016;5:1–18.
Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014;111:E2967–2976.
Matusek T, Gombos R, Szecsenyi A, Sanchez-Soriano N, Czibula A, Pataki C, et al. Formin proteins of the DAAM subfamily play a role during axon growth. J Neurosci. 2008;28:13310–9.
Kukulski W, Schorb M, Welsch S, Picco A, Kaksonen M, Briggs JA. Precise, correlated fluorescence microscopy and electron tomography of lowicryl sections using fluorescent fiducial markers. Methods Cell Biol. 2012;111:235–57.
Szikora S, Földi I, Tóth K, Migh E, Vig A, Bugyi B, et al. The formin DAAM is required for coordination of the actin and microtubule cytoskeleton in axonal growth cones. J Cell Sci. 2017;130:2506–19.
Lőrincz P, Tóth S, Benkő P, Lakatos Z, Boda A, Glatz G, et al. Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol. 2017;216:1937–47.
Gombos R, Migh E, Antal O, Mukherjee A, Jenny A, Mihály J. The formin DAAM functions as molecular effector of the planar cell polarity pathway during axonal development in Drosophila. J Neurosci. 2015;35:10154–67.
Tang HW, Liao HM, Peng WH, Lin HR, Chen CH, Chen GC. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev Cell. 2013;27:489–503.
Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, et al. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS ONE. 2012;7:e44214.
Lucas EP, Khanal I, Gaspar P, Fletcher GC, Polesello C, Tapon N, et al. The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J Cell Biol. 2013;201:875–85.
Gates J, Nowotarski SH, Yin H, Mahaffey JP, Bridges T, Herrera C, et al. Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev Biol. 2009;333:90–107.
Urwyler O, Cortinas-Elizondo F, Suter B. Drosophila sosie functions with beta(H)-Spectrin and actin organizers in cell migration, epithelial morphogenesis and cortical stability. Biol Open. 2012;1:994–1005.
Verheyen EM, Cooley L. Profilin mutations disrupt multiple actin-dependent processes during Drosophila development. Development. 1994;120:717–28.
Lambrechts A, Jonckheere V, Dewitte D, Vandekerckhove J, Ampe C. Mutational analysis of human profilin I reveals a second PI(4,5)-P2 binding site neighbouring the poly(L-proline) binding site. BMC Biochem. 2002;3:12.
McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–801.
Goncalves-Pimentel C, Gombos R, Mihaly J, Sanchez-Soriano N, Prokop A. Dissecting regulatory networks of filopodia formation in a Drosophila growth cone model. PLoS ONE. 2011;6:e18340.
Stavoe AK, Hill SE, Hall DH, Colon-Ramos DA. KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev Cell. 2016;38:171–85.
Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–9.
Nagy P, Szatmari Z, Sandor GO, Lippai M, Hegedus K, Juhasz G. Drosophila Atg16 promotes enteroendocrine cell differentiation via regulation of intestinal Slit/Robo signaling. Development. 2017;144:3990–4001.
Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600.
Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012;197:659–75.
Acknowledgements
We thank Szilvia Bozsó and Sarolta Pálfia for technical assistance and Adél Ürmösi for documentation of data, Zoltán Lipinszki for reagents, Anne Ephrussi for hosting VK in her group, and funders of our work: the Hungarian Academy of Sciences (Momentum LP2014/2 to GJ and PPD-222/2018 to PL), the National Research, Development, and Innovation Office (NKFIH) of Hungary (grants GINOP-2.3.2-15-2016-00032 to GJ, PV, and JM, GINOP-2.3.2-15-2016-00006 to GJ, PD128623 to SS and PD128357 to IF), the Hungarian Brain Research Program (2017-1.2.1-NKP-2017-00002 to JM) and an EMBO Short-Term Fellowship to VK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by H. Zhang
Rights and permissions
About this article
Cite this article
Kiss, V., Jipa, A., Varga, K. et al. Drosophila Atg9 regulates the actin cytoskeleton via interactions with profilin and Ena. Cell Death Differ 27, 1677–1692 (2020). https://doi.org/10.1038/s41418-019-0452-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-019-0452-0
This article is cited by
-
Identification and validation of autophagy-related genes in sepsis based on bioinformatics studies
Virology Journal (2025)
-
A neuroprotective role of Ufmylation through Atg9 in the aging brain of Drosophila
Cellular and Molecular Life Sciences (2023)
-
RNAseq and quantitative proteomic analysis of Dictyostelium knock-out cells lacking the core autophagy proteins ATG9 and/or ATG16
BMC Genomics (2021)
-
The autophagy protein ATG9A enables lipid mobilization from lipid droplets
Nature Communications (2021)