Abstract
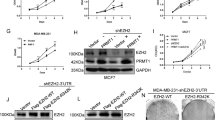
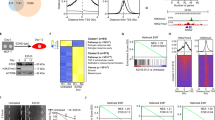
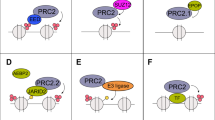
Enhancer of zeste homolog 2 (EZH2), a key histone methyltransferase and EMT inducer, is overexpressed in diverse carcinomas, including breast cancer. However, the molecular mechanisms of EZH2 dysregulation in cancers are still largely unknown. Here, we discover that EZH2 is asymmetrically dimethylated at R342 (meR342-EZH2) by PRMT1. meR342-EZH2 was found to inhibit the CDK1-mediated phosphorylation of EZH2 at T345 and T487, thereby attenuating EZH2 ubiquitylation mediated by the E3 ligase TRAF6. We also demonstrate that meR342-EZH2 resulted in a decrease in EZH2 target gene expression, but an increase in breast cancer cell EMT, invasion and metastasis. Moreover, we confirm the positive correlations among PRMT1, meR342-EZH2 and EZH2 expression in the breast cancer tissues. Finally, we report that high expression levels of meR342-EZH2 predict a poor clinical outcome in breast cancer patients. Our findings may provide a novel diagnostic target and promising therapeutic target for breast cancer metastasis.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun. 2015;6:6051.
Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100.
Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–83.
Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22:128–34.
Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24:59–71.
Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11.
Anwar T, Arellano-Garcia C, Ropa J, Chen YC, Kim HS, Yoon E, et al. p38-mediated phosphorylation at T367 induces EZH2 cytoplasmic localization to promote breast cancer metastasis. Nat Commun. 2018;9:2801.
Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci USA. 2014;111:1355–60.
Jin X, Yang C, Fan P, Xiao J, Zhang W, Zhan S, et al. CDK5/FBW7-dependent ubiquitination and degradation of EZH2 inhibits pancreatic cancer cell migration and invasion. J Biol Chem. 2017;292:6269–80.
Wan J, Zhan J, Li S, Ma J, Xu W, Liu C, et al. PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res. 2015;43:3591–604.
Wan L, Xu K, Wei Y, Zhang J, Han T, Fry C, et al. Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function. Mol Cell. 2018;69:279–91.e275.
Wu SC, Zhang Y. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem. 2011;286:28511–9.
Lo PW, Shie JJ, Chen CH, Wu CY, Hsu TL, Wong CH. O-GlcNAcylation regulates the stability and enzymatic activity of the histone methyltransferase EZH2. Proc Natl Acad Sci USA. 2018;115:7302–07.
Jarrold J, Davies CC. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol Med. 2019;25:993–1009.
Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer 2013;13:37–50.
Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24.
Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13.
Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, et al. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–30.
Cho JH, Lee MK, Yoon KW, Lee J, Cho SG, Choi EJ. Arginine methylation-dependent regulation of ASK1 signaling by PRMT1. Cell Death Differ. 2012;19:859–70.
Davies CC, Chakraborty A, Diefenbacher ME, Skehel M, Behrens A. Arginine methylation of the c-Jun coactivator RACO-1 is required for c-Jun/AP-1 activation. EMBO J. 2013;32:1556–67.
Sakamaki J, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci USA. 2011;108:6085–90.
Wang Y, Hsu JM, Kang Y, Wei Y, Lee PC, Chang SJ, et al. Oncogenic functions of Gli1 in pancreatic adenocarcinoma are supported by its PRMT1-mediated methylation. Cancer Res. 2016;76:7049–58.
Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–31.
Yang JH, Chiou YY, Fu SL, Shih IY, Weng TH, Lin WJ, et al. Arginine methylation of hnRNPK negatively modulates apoptosis upon DNA damage through local regulation of phosphorylation. Nucleic Acids Res. 2014;42:9908–24.
Cha B, Kim W, Kim YK, Hwang BN, Park SY, Yoon JW, et al. Methylation by protein arginine methyltransferase 1 increases stability of Axin, a negative regulator of Wnt signaling. Oncogene. 2011;30:2379–89.
Bai J, Wu K, Cao MH, Yang Y, Pan Y, Liu H, et al. SCF(FBXO22) targets HDM2 for degradation and modulates breast cancer cell invasion and metastasis. Proc Natl Acad Sci USA. 2019;116:11754–63.
Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316.
Coutsias EA, Seok C, Jacobson MP, Dill KA. A kinematic view of loop closure. J Comput Chem. 2004;25:510–28.
Huang J, MacKerell AD. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem. 2013;34:2135–45.
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802.
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–93.
Yu J, Qin B, Moyer AM, Nowsheen S, Liu T, Qin S, et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest. 2018;128:2376–88.
Liu LM, Sun WZ, Fan XZ, Xu YL, Cheng MB, Zhang Y. Methylation of C/EBPalpha by PRMT1 inhibits its tumor-suppressive function in breast cancer. Cancer Res. 2019;79:2865–77.
Wooderchak WL, Zang T, Zhou ZS, Acuna M, Tahara SM, Hevel JM. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry. 2008;47:9456–66.
Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, et al. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest. 2015;125:4529–43.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (81802637, 81672845 and 81872304); the Natural Science Foundation of Jiangsu Province (BK20180989); the National Postdoctoral Research Funds (2019M651971); the Natural Science Foundation of Jiangsu Province Universities (18KJB310016); the Research Foundation of Xuzhou Medical University (D2018020); the Xuzhou City Science and Technology Plan Project (KC19065); the Jiangsu Provincial Key Medical Discipline, the Project of Invigorating Health Care through Science, Technology and Education (ZDXKA2016014).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by A. Oberst
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Z., Wang, D., Lu, J. et al. Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ 27, 3226–3242 (2020). https://doi.org/10.1038/s41418-020-00615-9
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-020-00615-9
This article is cited by
-
S-nitrosylation of EZH2 alters PRC2 assembly, methyltransferase activity, and EZH2 stability to maintain endothelial homeostasis
Nature Communications (2025)
-
TRIM21-mediated PRMT1 degradation attenuates colorectal cancer malignant progression
Cell Death & Disease (2025)
-
PRMT1-catalyzed NUSAP1 methylation enhances Notch2 signaling and 5-FU resistance in gastric cancer
Cell Death & Disease (2025)
-
Enhancer zeste homolog 2 (EZH2) targeting by small interfering RNA (siRNA); recent advances and prospect
Naunyn-Schmiedeberg's Archives of Pharmacology (2025)
-
A coagulation-related long non-coding RNA signature to predict prognosis and immune features of breast cancer
Discover Oncology (2025)