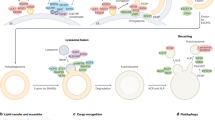
Abstract
Macroautophagy (autophagy) delivers intracellular constituents to the lysosome to promote catabolism. During development in multiple organisms, autophagy mediates various cellular processes, including survival during starvation, programmed cell death, phagocytosis, organelle elimination, and miRNA regulation. Our current understanding of autophagy has been enhanced by developmental biology research during the last quarter of a century. Through experiments that focus on animal development, fundamental mechanisms that control autophagy and that contribute to disease were elucidated. Studies in embryos revealed specific autophagy molecules that mediate the removal of paternally derived mitochondria, and identified autophagy components that clear protein aggregates during development. Importantly, defects in mtDNA inheritance, or removal of paternal mtDNA via mitochondrial autophagy, can contribute to mitochondrial-associated disease. In addition, impairment of the clearance of protein aggregates by autophagy underlies neurodegenerative diseases. Experiments in multiple organisms also reveal conserved mechanisms of tissue remodeling that rely on the cooperation between autophagy and apoptosis to clear cell corpses, and defects in autophagy and apoptotic cell clearance can contribute to inflammation and autoimmunity. Here we provide an overview of key developmental processes that are mediated by autophagy in multiple animals.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies: Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604–17.
Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–96.
Mony VK, Benjamin S, O’Rourke EJ. A lysosome-centered view of nutrient homeostasis. Autophagy. 2016;12:619–31.
Lawrence RE, Zoncu R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat Cell Biol. 2019;21:133–42.
de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–92.
Santambrogio L, Cuervo AM. Chasing the elusive mammalian microautophagy. Autophagy. 2011. https://doi.org/10.4161/auto.7.6.15287.
Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodaton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–5.
Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–3.
Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275:27447–56.
Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–34.
Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202.
Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol. 1968;53:687–733.
Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11.
Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–80.
Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74.
Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6.
Yang Z, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31.
Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77.
Mercer TJ, Gubas A, Tooze SA. A molecular perspective of mammalian autophagosome biogenesis. J Biol Chem. 2018;293:5386–95.
Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305.
Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013;55:39–50.
Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–76.
Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–76.
Reggiori F, Ungermann C. Autophagosome maturation and fusion. J Mol Biol. 2017;429:486–96.
Rong Y, McPhee CK, McPhee C, Deng S, Huang L, Chen L, et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci USA. 2011;108:7826–31.
Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–15.
Song WH, Yi YJ, Sutovsky M, Meyers S, Sutovsky P. The ART and science of sperm mitophagy. Autophagy. 2016;12:2510–1.
Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–4.
Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–7.
Shakes DC, Ward S. Mutations that disrupt the morphogenesis and localization of a sperm-specific organelle in Caenorhabditis elegans. Dev Biol. 1989;231:1–23.
Wei Y, Chiang WC, Sumpter R, Mishra P, Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–38.
Hird SN, Paulsen JE, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–12.
Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, et al. SEPA-1 mediates the specific recognition and degradation of p granule components by autophagy in C. elegans. Cell. 2009;136:308–21.
Komatsu M, Waguri S, Koike M, Sou Y-S, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63.
Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, et al. Ref(2)P, the drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–71.
Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–55.
Zhang G, Wang Z, Du Z, Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174:1492–.e22.
Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–20.
Merz EA, Brinster RL, Brunner S, Chen HY. Degradation during preimplantation development. J Reprod Fert. 1981;61:451–418.
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6.
Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34.
Yamamoto A, Mizushima N, Tsukamoto S. Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol Reprod. 2014;91:1–7.
Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc Natl Acad Sci USA. 2011;108:11121–6.
Feng Y, Kang HH, Wong PM, Gao M, Wang P, Jiang X. Unc-51-like kinase (ULK) complex-independent autophagy induced by hypoxia. Protein Cell. 2019;10:376–81.
Corona Velazquez A, Corona AK, Klein KA, Jackson WT. Poliovirus induces autophagic signaling independent of the ULK1 complex. Autophagy. 2018;14:1201–13.
Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–82.
Maria Fimia G, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5.
Yazdankhah M, Farioli-Vecchioli S, Tonchev AB, Stoykova A, Cecconi F. The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis. 2014;5:e1403.
Cianfanelli V, De Zio D, Di Bartolomeo S, Nazio F, Strappazzon F, Cecconi F. Ambra1 at a glance. J Cell Sci. 2015;128:2003–8.
Grimsley C, Ravichandran KS. Cues for apoptotic cell engulfment: eat-me, don’t eat-me and come-get-me signals. Trends Cell Biol. 2003;13:648–56.
Conradt B, Wu YC, Xue D. Programmed cell death during Caenorhabditis elegans development. Genetics. 2016;203:1533–62.
Huang S, Jia K, Wang Y, Zhou Z, Levine B. Autophagy genes function in apoptotic cell corpse clearance during C. Elegans embryonic development. Autophagy. 2013;9:1–12.
Cheng S, Wu Y, Lu Q, Yan J, Zhang H, Wang X. Autophagy genes coordinate with the class II PI/PtdIns 3-kinase PIKI-1 to regulate apoptotic cell clearance in C. elegans. Autophagy. 2013;9:2022–32.
Jenzer C, Simionato E, Largeau C, Scarcelli V, Lefebvre C, Legouis R. Autophagy mediates phosphatidylserine exposure and phagosome degradation during apoptosis through specific functions of GABARAP/LGG-1 and LC3/LGG-2. Autophagy. 2019;15:228–41.
Li W, Zou W, Yang Y, Chai Y, Chen B, Cheng S, et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol. 2012;197:27–35.
Ruck A, Attonito J, Garces KT, Núnez L, Palmisano NJ, Rubel Z, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011;7:386–400.
Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Inactivation of the autophagy Gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–7.
Reed BH, Wilk R, Schöck F, Lipshitz HD. Integrin-dependent apposition of Drosophila extraembryonic membranes promotes morphogenesis and prevents anoikis. Curr Biol. 2004;14:372–80.
Mohseni N, McMillan SC, Chaudhary R, Mok J, Reed BH. Autophagy promotes caspase-dependent cell death during Drosophila development. Autophagy. 2009;5:329–38.
Cormier O, Mohseni N, Voytyuk I, Reed BH. Autophagy can promote but is not required for epithelial cell extrusion in the amnioserosa of the Drosophila embryo. Autophagy. 2012;8:252–64.
Coucouvanis E, Martin GR. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 1995;83. https://doi.org/10.1016/0092-8674(95)90169-8.
Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–46.
Mellén MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15:1279–90.
Mellén MA, De La Rosa EJ, Boya P. Autophagy is not universally required for phosphatidyl-serine exposure and apoptotic cell engulfment during neural development. Autophagy. 2009;5:964–72.
Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91.
Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55.
Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–6.
Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–7.
Lehmann R, Nüsslein-Volhard C. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev Biol. 1987;119:402–17.
Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–49.
Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205:240–53.
Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–14.
Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437–47.
Zhang L, Ding L, Cheung TH, Dong MQ, Chen J, Sewell AK, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their Interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613.
Zhang P, Zhang H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 2013;14:568–76.
Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–21.
Gozuacik D, Akkoc Y, Gulfem Ozturk D, Kocak M. Autophagy-regulating microRNAs and cancer. Front Oncol. 2017;7:1–22.
Petri R, Pircs K, Jönsson ME, Åkerblom M, Brattås PL, Klussendorf T, et al. let‐7 regulates radial migration of new‐born neurons through positive regulation of autophagy. EMBO J. 2017;36:1379–91.
Sou YS, Waguri S, Iwata JI, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75.
Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–6.
Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–8.
Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2012;493:679.
Schin KS, Clever U. Lysosomal and free acid phosphatase in salivary glands of chironomus tentans. Science. 1965;150:1053–5.
Locke M, Collins JV. The structure and formation of protein granules in the fat body of an insect. J Cell Biol. 1965;26:857–84.
Scharrer B. Ultrastructural study of the regressing prothoracic glands of blattarian insects. Z für Zellforsch und Mikroskopische Anat. 1966;69:1–21.
Schin K, Laufer H. Studies of programmed salivary gland regression during larval-pupal transformation in Chironomus thummi. Exp Cell Res. 1973;82:335–40.
Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Antheraea polyphemus and Manduca sexta (Insecta, lepidoptera) with particular reference to cellular autophagy. J Morphol. 1977;154:39–57.
Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–55.
Baehrecke EH. Autophagic programmed cell death in Drosophila. Cell Death Differ. 2003;10:940–5.
Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in drosophila. Curr Biol. 2009;19:1741–6.
Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in drosophila. Cell. 2007;131:1137–48.
Rusten TE, Lindmo K, Juhász G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K Pathway. Dev Cell. 2004;7:179–92.
Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003;13:350–7.
Lee C-Y, Cooksey BAK, Baehrecke EH. Steroid regulation of midgut cell death during drosophila development. Dev Biol. 2002;250:101–11.
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13.
Velentzas PD, Zhang L, Das G, Chang TK, Nelson C, Kobertz WR, et al. The proton-coupled monocarboxylate transporter hermes is necessary for autophagy during cell death. Dev Cell. 2018;47:281–93.
Nelson C, Ambros V, Baehrecke EH. miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Mol Cell. 2014;56:376–88.
Martin DN, Baehrecke EH. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–84.
Lin L, Rodrigues FSLM, Kary C, Contet A, Logan M, Baxter RHG, et al. Complement-related regulates autophagy in neighboring cells. Cell. 2017;170:158–.e8.
McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–7.
Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in drosophila. Curr Biol. 2009. https://doi.org/10.1016/j.cub.2009.08.042.
Chang T-K, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J, et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013;15:1067–78.
Anding AL, Wang C, Chang TK, Sliter DA, Powers CM, Hofmann K, et al. Vps13D encodes a ubiquitin-binding protein that is required for the regulation of mitochondrial size and clearance. Curr Biol. 2018;28:287–95.
Felix DA, Gutiérrez-Gutiérrez Ó, Espada L, Thems A, González-Estévez C. It is not all about regeneration: planarians striking power to stand starvation. Semin Cell Dev Biol. 2019;87:169–81.
Bowen ID, Ryder TA. Cell autolysis and deletion in the planarian polycelis tenuis Iijima. Cell Tissue Res. 1974;154:265–74.
Bowen ID, Ryder TA, Dark C. The effects of starvation on the planarian worm Polycelis tenuis iijima. Cell Tissue Res. 1976;169:193–209.
Bowen ID, den Hollander JE, Lewis GHJ. Cell death and acid phosphatase activity in the regenerating planarian polycelis tenuis Iijima. Differentiation. 1982;21:160–7.
Tettamanti G, Salo E, Gonzalez-Estevez C, Felix D, Grimaldi A, Eguileor M. Autophagy in invertebrates: insights into development, regeneration and body remodeling. Curr Pharm Des. 2008;14:116–25.
González-Estévez C, Felix DA, Smith MD, Paps J, Morley SJ, James V, et al. SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 2012;8:e1002619.
Kang J, Dong Z, Hao Q, Wang J, Chen G, Liu D. The regulation of rapamycin in planarian Dugesia japonica Ichikawa & T Kawakatsu, 1964 regeneration according to TOR signaling pathway. Ecotoxicol Environ Saf. 2019;185:109680.
Baguñá J, Romero R. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia. 1981;84:181–94.
González-Estévez C, Felix DA, Rodríguez-Esteban G, Aziz Aboobaker A. Decreased neoblast progeny and increased cell death during starvation-induced planarian degrowth. Int J Dev Biol. 2012;56:83–91.
González-Estévez C, Felix DA, Aboobaker AA, Saló E. Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation. Proc Natl Acad Sci USA. 2007;104:13373–8.
Yuan J, Wang Z, Zou D, Peng Q, Peng R, Zou F. Expression profiling of planarians shed light on a dual role of programmed cell death during the regeneration. J Cell Biochem. 2018;119:5875–84.
Pfefferli C, Jaźwińska A. The art of fin regeneration in zebrafish. Regeneration. 2015;2:72–83.
Varga M, Sass M, Papp D, Takács-Vellai K, Kobolak J, Dinnyés A, et al. Autophagy is required for zebrafish caudal fin regeneration. Cell Death Differ. 2014;21:547–56.
Saera-Vila A, Kasprick DS, Junttila TL, Grzegorski SJ, Louie KW, Chiari EF, et al. Myocyte dedifferentiation drives extraocular muscle regeneration in adult zebrafish. Investig Ophthalmol Vis Sci. 2015;56:4977–93.
Saera-Vila A, Kish PE, Louie KW, Grzegorski SJ, Klionsky DJ, Kahana A. Autophagy regulates cytoplasmic remodeling during cell reprogramming in a zebrafish model of muscle regeneration. Autophagy. 2016;12:1864–75.
Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci USA. 2012;109:21528–33.
Biteau B, Karpac J, Supoyo S, DeGennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:e1001159.
Jiang H, Edgar BA. Intestinal stem cell function in Drosophila and mice. Curr Opin Genet Dev. 2012;22:354–60.
Nagy P, Sándor GO, Juhász G. Autophagy maintains stem cells and intestinal homeostasis in Drosophila. Sci Rep. 2018;8:4644.
Zhang P, Holowatyj AN, Roy T, Pronovost SM, Marchetti M, Liu H, et al. An SH3PX1-dependent endocytosis-autophagy network restrains intestinal stem cell proliferation by counteracting EGFR-ERK signaling. Dev Cell. 2019;49:574–89.
Tang HW, Liao HM, Peng WH, Lin HR, Chen CH, Chen GC. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev Cell. 2013;27:489–503.
Elliott MR, Ravichandran KS. Clearance of apoptotic cells: Implications in health and disease. J Cell Biol. 2010;189:1059–70.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by F. Pentimalli
Rights and permissions
About this article
Cite this article
Allen, E.A., Baehrecke, E.H. Autophagy in animal development. Cell Death Differ 27, 903–918 (2020). https://doi.org/10.1038/s41418-020-0497-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-020-0497-0
This article is cited by
-
The proteostatic landscape of healthy human oocytes
The EMBO Journal (2025)
-
Autophagic activity in the midgut cells of three arachnids responds selectively to different modes of overwintering in caves
Protoplasma (2025)
-
Apolipoprotein B100 acts as a tumor suppressor in ovarian cancer via lipid/ER stress axis-induced blockade of autophagy
Acta Pharmacologica Sinica (2025)
-
Homocysteine and mitochondrial quality control in diabetic retinopathy
Eye and Vision (2024)
-
Plasma SQSTM1/p62 act as a biomarker for steroid-induced osteonecrosis of the femoral head
Scientific Reports (2024)