Abstract
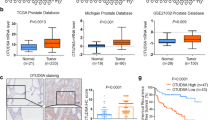
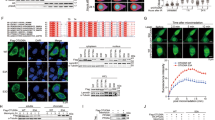
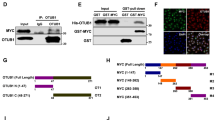
MYC drives the tumorigenesis of human cancers, including prostate cancer (PrCa), thus deubiquitinase (DUB) that maintains high level of c-Myc oncoprotein is a rational therapeutic target. Several ubiquitin-specific protease (USP) family members of DUB have been reported to deubiquitinate c-Myc, but none of them is the physiological DUB for c-Myc in PrCa. By screening all the DUBs, here we reveal that OTUD6A is exclusively amplified and overexpressed in PrCa but not in other cancers, eliciting a prostatic-specific oncogenic role through deubiquitinating and stabilizing c-Myc oncoprotein. Moreover, genetic ablation of OTUD6A efficiently represses prostatic tumorigenesis of both human PrCa cells and the Hi-Myc transgenic PrCa mice, via reversing the metabolic remodeling caused by c-Myc overexpression in PrCa. These results indicate that OTUD6A is a physiological DUB for c-Myc in PrCa setting and specifically promotes prostatic tumorigenesis through stabilizing c-Myc oncoprotein, suggesting that OTUD6A could be a unique therapeutic target for Myc-driven PrCa.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. RNA-seq data discussed in this paper have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE189716.
Change history
19 April 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41418-022-00978-1
References
Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and prostate cancer. Genes Cancer. 2010;1:617–28.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–73.
Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8.
Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51.
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9.
Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9.
Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60.
Ribeiro FR, Jeronimo C, Henrique R, Fonseca D, Oliveira J, Lothe RA, et al. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–70.
Ribeiro FR, Henrique R, Martins AT, Jeronimo C, Teixeira MR. Relative copy number gain of MYC in diagnostic needle biopsies is an independent prognostic factor for prostate cancer patients. Eur Urol. 2007;52:116–25.
Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, et al. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol. 2013;44:1617–23.
Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–6.
Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–9.
Dhanasekaran SM, Dash A, Yu J, Maine IP, Laxman B, Tomlins SA, et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–5.
Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–67.
Gil J, Kerai P, Lleonart M, Bernard D, Cigudosa JC, Peters G, et al. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005;65:2179–85.
Williams K, Fernandez S, Stien X, Ishii K, Love HD, Lau YF, et al. Unopposed c-MYC expression in benign prostatic epithelium causes a cancer phenotype. Prostate. 2005;63:369–84.
Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38.
Kim J, Eltoum I-EA, Roh M, Wang J, Abdulkadir SA. Interactions between cells with distinct mutations in c-MYC and Pten in prostate cancer. PLoS Genet. 2009;5:e1000542.
Ellis L, Ku S, Li Q, Azabdaftari G, Seliski J, Olson B, et al. Generation of a C57BL/6 MYC-driven mouse model and cell line of prostate cancer. Prostate. 2016;76:1192–202.
Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer cell. 2019;36:483–97 e415.
Clausen DM, Guo J, Parise RA, Beumer JH, Egorin MJ, Lazo JS, et al. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J Pharm Exp Ther. 2010;335:715–27.
Allen-Petersen BL, Sears RC. Mission possible: advances in MYC therapeutic targeting in cancer. BioDrugs. 2019;33:539–53.
Leonard D, Huang W, Izadmehr S, O’Connor CM, Wiredja DD, Wang Z, et al. Selective PP2A enhancement through biased heterotrimer stabilization. Cell. 2020;181:688–701 e616.
Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26:2832–44.
Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25.
Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–90.
Geng C, Kaochar S, Li M, Rajapakshe K, Fiskus W, Dong J, et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36:4767–77.
Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ, et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232:3664–76.
Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–74.
Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2015;112:3734–9.
Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–67.
Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17:1169–81.
Liu J, Peng Y, Shi L, Wan L, Inuzuka H, Long J, et al. Skp2 dictates cell cycle-dependent metabolic oscillation between glycolysis and TCA cycle. Cell Res. 2021;31:80–93.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disco. 2012;2:401–4.
Wang Z, Song Y, Ye M, Dai X, Zhu X, Wei W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol. 2020;17:339–50.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9.
Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–84.
Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83.
Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18.
Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–79.
Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14.
Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35.
Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Disco. 2015;5:1024–39.
Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93.
Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–63.
Singh KB, Hahm ER, Kim SH, Wendell SG, Singh SV. A novel metabolic function of Myc in regulation of fatty acid synthesis in prostate cancer. Oncogene. 2021;40:592–602.
Devi GR, Beer TM, Corless CL, Arora V, Weller DL, Iversen PL. In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin Cancer Res. 2005;11:3930–8.
Iversen PL, Arora V, Acker AJ, Mason DH, Devi GR. Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin Cancer Res. 2003;9:2510–9.
Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17.
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113:7124–9.
Acknowledgements
We thank Dr. Zhihui Feng, Dr. Jian Ding, and Dr. Yongping Shao for suggestions and comments on this work. We thank Dr. Yong Yang from China Pharmaceutical University for his kind help for providing Hi-Myc mice as gift.
Funding
This work was supported by the National Natural Science Foundation of China [81802787, 31700684, 31870848, 31770917 and 32171102], the National Basic Research Program of China [973 Program 2015CB856302 and 2015CB553602], the China Postdoctoral Science Foundation [2018M643673], and the National Key Research and Development Projects of China [2019YFD1002400].
Author information
Authors and Affiliations
Contributions
The idea was conceived by WW, LZ, JKL, and JGL. YP and JL designed and performed most of the experiments with assistance from ZW, CC, TZ, PG, ZH, HL, YW, JG, and JZ. YP and JL wrote the manuscript. WW, LZ, JKL, and JGL supervised the study and edited the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All animal experiments were approved by the Animal Use and Care Committee of the School of Life Science and Technology, Xi’an Jiaotong University, based on the Guide for the Care and Use of Laboratory Animals: eighth edition (ISBN10: 0-309-15396-4).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: Due to a figure error.
Edited by A Degterev
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peng, Y., Liu, J., Wang, Z. et al. Prostate-specific oncogene OTUD6A promotes prostatic tumorigenesis via deubiquitinating and stabilizing c-Myc. Cell Death Differ 29, 1730–1743 (2022). https://doi.org/10.1038/s41418-022-00960-x
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-022-00960-x
This article is cited by
-
Unveiling the molecular profile of a prostate carcinoma: implications for personalized medicine
Biology Direct (2024)
-
Deubiquitination of CDC6 by OTUD6A promotes tumour progression and chemoresistance
Molecular Cancer (2024)
-
MYC induces CDK4/6 inhibitors resistance by promoting pRB1 degradation
Nature Communications (2024)
-
Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance
Cancer and Metastasis Reviews (2024)
-
Protein degradation: expanding the toolbox to restrain cancer drug resistance
Journal of Hematology & Oncology (2023)