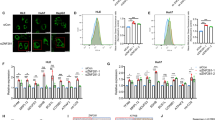
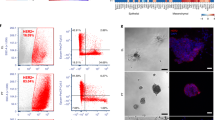
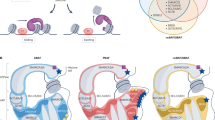
Abstract
Although the Hedgehog (Hh) pathway plays an evolutionarily conserved role from Drosophila to mammals, some divergences also exist. Loss of Sufu, an important component of the Hh pathway, does not lead to an obvious developmental defect in Drosophila. However, in mammals, loss of SUFU results in serious disorder, even various cancers. This divergence suggests that SUFU plays additional roles in mammalian cells, besides regulating the Hh pathway. Here, we identify that the transcription factor ZNF281 is a novel binding partner of SUFU. Intriguingly, the Drosophila genome does not encode any homologs of ZNF281. SUFU is able to suppress ZNF281-induced tumor cell migration and DNA damage repair by inhibiting ZNF281 activity. Mechanistically, SUFU binds ZNF281 to mask the nuclear localization signal of ZNF281, culminating in ZNF281 cytoplasmic retention. In addition, SUFU also hampers the interactions between ZNF281 and promoters of target genes. Finally, we show that SUFU is able to inhibit ZNF281-induced tumor cell migration using an in vivo model. Taken together, these results uncover a Hh-independent mechanism of SUFU exerting the anti-tumor role, in which SUFU suppresses tumor cell migration through antagonizing ZNF281. Therefore, this study provides a possible explanation for the functional divergence of SUFU in mammals and Drosophila.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
All relevant data are available from the corresponding author upon reasonable request.
References
Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–12.
Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–86.
Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406.
Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–29.
Kogerman P, Grimm T, Kogerman L, Krause D, Undén AB, Sandstedt§ B, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1:312–9.
Pearse VR,II, Collier LS, Scott MP, Tabin CJ. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev Biol. 1999;212:323–36.
Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8:583–6.
Me´thot N KB. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 1999;96:819–31.
Han YH, Shi Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci USA. 2015;112:6383–8.
Zhang Q, Zhang L, Wang B, Ou C-Y, Chien C-T, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–29.
Préat T. Characterization of Suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics 1992;132:725–36.
Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–97.
Ding Q, Fukami S-I, Meng X, Nishizaki§ Y, Zhang X, Sasaki§ H, et al. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–22.
Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–82.
Shi Q, Han Y, Jiang J. Suppressor of fused impedes Ci/Gli nuclear import by opposing Trn/Kapbeta2 in Hedgehog signaling. J Cell Sci. 2014;127:1092–103.
Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–10.
Law DJ, Du M, Law GL, Merchant JL. ZBP-99 defines a conserved family of transcription factors and regulates ornithine decarboxylase gene expression. Biochem Biophys Res Commun. 1999;262:113–20.
Lisowskya T, Polosab PL, Saglianob A, Robertib M, Gadaletab MN, Cantatoreb P. Identification of human GC-box-binding zinc finger protein, a new Krüppel-like zinc finger protein, by the yeast one-hybrid screening with a GC-rich target sequence. FEBS Lett. 1999;45:369–74.
Fidalgo M, Huang X, Guallar D, Sanchez-Priego C, Valdes VJ, Saunders A, et al. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell. 2016;19:355–69.
Ishiuchi T, Ohishi H, Sato T, Kamimura S, Yorino M, Abe S, et al. Zfp281 shapes the transcriptome of trophoblast stem cells and is essential for placental development. Cell Rep. 2019;27:1742–54.e6
Pieraccioli M, Nicolai S, Antonov A, Somers J, Malewicz M, Melino G, et al. ZNF281 contributes to the DNA damage response by controlling the expression of XRCC2 and XRCC4. Oncogene 2016;35:2592–601.
Xue Y, Ding M, Xue L, Luo J. CircAGFG1 sponges miR-203 to promote EMT and metastasis of non-small-cell lung cancer by upregulating ZNF281 expression. Thorac Cancer. 2019;10:1692–701.
Ji W, Mu Q, Liu XY, Cao XC, Yu Y. ZNF281-miR-543 feedback loop regulates transforming growth factor-beta-induced breast cancer metastasis. Mol Ther Nucleic Acids. 2020;21:98–107.
Starzynska A, Sobocki BK, Sejda A, Sakowicz-Burkiewicz M, Szot O, Jereczek-Fossa BA. ZNF-281 as the potential diagnostic marker of oral squamous cell carcinoma. Cancers (Basel). 2021;13:2661.
Hahn S, Jackstadt R, Siemens H, Hunten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–95.
Zhu Y, Zhou Q, Zhu G, Xing Y, Li S, Ren N, et al. GSK-3β phosphorylation-dependent degradation of ZNF281 by β-TrCP2 suppresses colorectal cancer progression. Oncotarget 2017;8:88599–612.
Hahn S, Hermeking H. ZNF281/ZBP-99: a new player in epithelial-mesenchymal transition, stemness, and cancer. J Mol Med. 2014;92:571–81.
Peng Y, Qin Y, Zhang X, Deng S, Yuan Y, Feng X, et al. MiRNA-20b/SUFU/Wnt axis accelerates gastric cancer cell proliferation, migration and EMT. Heliyon 2021;7:e06695.
Peng Y, Zhang X, Lin H, Deng S, Qin Y, He J, et al. Dual activation of Hedgehog and Wnt/beta-catenin signaling pathway caused by downregulation of SUFU targeted by miRNA-150 in human gastric cancer. Aging (Albany NY). 2021;13:10749–69.
Peng Y, Zhang X, Lin H, Deng S, Qin Y, Yuan Y, et al. SUFU mediates EMT and Wnt/beta-catenin signaling pathway activation promoted by miRNA-324-5p in human gastric cancer. Cell Cycle. 2020;19:2720–33.
Zhang Z, Zou Y, Liang M, Chen Y, Luo Y, Yang B, et al. Suppressor of fused (Sufu) promotes epithelial-mesenchymal transition (EMT) in cervical squamous cell carcinoma. Oncotarget 2017;8:114226–38.
Liu A. Proteostasis in the Hedgehog signaling pathway. Semin Cell Dev Biol. 2019;93:153–63.
Nano M, Gemble S, Simon A, Pennetier C, Fraisier V, Marthiens V, et al. Cell-cycle asynchrony generates DNA damage at mitotic entry in polyploid cells. Curr Biol. 2019;29:3937–45.
Mladenov E, Magin S, Soni A, G I. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: cell cycle and proliferation-dependent regulation. Semin Cancer Biol. 2016;37-38:51–64.
Roos PW, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–50.
Miles WO, Dyson NJ, Walker JA. Modeling tumor invasion and metastasis in Drosophila. Dis Model Mech. 2011;4:753–61.
Niisslein-Volhard C EW. Mutations affecting segment number and polarity in Drosophila. Nature 1980;287:795–801.
Heemskerk J SD. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76:449–60.
Chi S, Huang S, Li C, Zhang X, He N, Bhutani MS, et al. Activation of the hedgehog pathway in a subset of lung cancers. Cancer Lett. 2019;244:53–60.
Sheng T, Li C, Zhang X, Chi S, He N, Chen K, et al. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29.
Guerrini-Rousseau L, Dufour C, Varlet P, Masliah-Planchon J, Bourdeaut F, Guillaud-Bataille M, et al. Germline SUFU mutation carriers and medulloblastoma: clinical characteristics, cancer risk, and prognosis. Neuro Oncol. 2018;20:1122–32.
Xu F, Li H, C H. LIFR-AS1 modulates Sufu to inhibit cell proliferation and migration by miR-197-3p in breast cancer. Biosci Rep. 2019;39:BSR20180551.
Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol mutagenesis. 2017;58:235–63.
O’Connor MJ. Targeting the DNA damage response in cancer. Mol cell. 2015;60:547–60.
Zhou F, Huang D, Li Y, Hu G, Rao H, Q L, et al. Nek2A/SuFu feedback loop regulates Gli-mediated Hedgehog signaling pathway. Int J Oncol. 2017;50:373–80.
Sun X, Ding Y, Zhan M, Li Y, Gao D, Wang G, et al. Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat Commun. 2019;10:411.
Ding Y, Wang G, Zhan M, Sun X, Deng Y, Zhao Y, et al. Hippo signaling suppresses tumor cell metastasis via a Yki-Src42A positive feedback loop. Cell Death Dis. 2021;12:1126.
Zhou Z, Xu C, Chen P, Liu C, Pang S, Yao X, et al. Stability of HIB-Cul3 E3 ligase adaptor HIB is regulated by self-degradation and availability of its substrates. Sci Rep. 2015;5:12709.
Zhou Z, Yao X, Li S, Xiong Y, Dong X, Zhao Y, et al. Deubiquitination of Ci/Gli by Usp7/HAUSP regulates hedgehog signaling. Dev Cell. 2015;34:58–72.
Zhan M, Sun X, Liu J, Li Y, Li Y, He X, et al. Usp7 promotes medulloblastoma cell survival and metastasis by activating Shh pathway. Biochem Biophys Res Commun. 2017;484:429–34.
Acknowledgements
We sincerely thank Prof. Lei Xue (Tongji University, China) for providing scrib-RNAi fly and Prof. Yong Li (Nanchang University, China) for providing lentiviral vectors.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31922011, 31802012, and ZR2021MH137), the National Key Research and Development Program of China (2017YFA0205200), Program for Scientific Research Innovation Team of Young Scholar in Colleges and Universities of Shandong Province (2019KJE009), and the Construction Engineering Special Fund of “Taishan Scholars” (no. Ts201712022).
Author information
Authors and Affiliations
Contributions
MZ, ZZ, and JD designed this study and provide financial support. YD, DP, JX, YZ, WD, SY, and LS carried out the experiments and analyzed the data. SY and LS provided HCC cell lines. YD and DP performed mouse experiments. YZ analyzed cell migration in Drosophila. YD and ZZ wrote the article with the assistance of all authors. MZ, ZZ, and JD supervised this study and modified the manuscript. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All animal experiments were approved by the Institutional Animal Care Committee of China Pharmaceutical University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by G. Del Sal
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, Y., Peng, D., Xiao, J. et al. Inhibition of the transcription factor ZNF281 by SUFU to suppress tumor cell migration. Cell Death Differ 30, 702–715 (2023). https://doi.org/10.1038/s41418-022-01073-1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-022-01073-1
This article is cited by
-
E3 ligase TRIM65 alleviates intestinal ischemia/reperfusion injury through inhibition of TOX4-mediated apoptosis
Cell Death & Disease (2024)
-
tRF3-IleAAT reduced extracellular matrix synthesis in diabetic kidney disease mice by targeting ZNF281 and inhibiting ferroptosis
Acta Pharmacologica Sinica (2024)
-
TRIM65 promotes renal cell carcinoma through ubiquitination and degradation of BTG3
Cell Death & Disease (2024)
-
Identification and validation of the role of ZNF281 in 5-fluorouracil chemotherapy of gastric cancer
Journal of Cancer Research and Clinical Oncology (2024)
-
Deep dissection of stemness-related hierarchies in hepatocellular carcinoma
Journal of Translational Medicine (2023)