Abstract
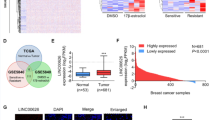
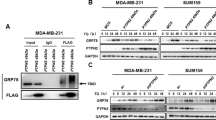
Clinical treatment options for triple-negative breast cancer (TNBC) are currently limited to chemotherapy because of a lack of effective therapeutic targets. Recent evidence suggests that long noncoding RNAs (lncRNAs) encode bioactive peptides or proteins, thereby playing noncanonical yet significant roles in regulating cellular processes. However, the potential of lncRNA-translated products in cancer progression remains largely unknown. In this study, we identified a previously undocumented small protein encoded by the lncRNA LINC02870. This protein is localized at the endoplasmic reticulum (ER) and participates in ER stress, thus, we named it the endoplasmic reticulum stress protein (ERSP). ERSP was highly expressed in TNBC tissues, and elevated LINC02870 content was correlated with poor prognosis in TNBC patients. Loss of ERSP inhibited TNBC growth and metastasis both in vitro and in vivo. The pro-oncogenic effects of ERSP could be attributed to its selective activation of the IRE1α/XBP1s branch. ERSP enhances the unfolded protein response (UPR) by interacting with XBP1s, facilitating the nuclear accumulation of XBP1s, thereby promoting the expression of ER stress-related genes. These findings highlight the regulatory role of the lncRNA-encoded protein ERSP in ER stress and suggest that it is a potential therapeutic target for TNBC.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq data are available at the National Genomics Data Center database: https://github.com/Wangjunjian-lab/Small-protein-ERSP-encoded-by-LINC02870. The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the iProX partner repository (https://www.iprox.cn) with the dataset identifier PXD048235.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Asleh K, Riaz N, Nielsen TO. Heterogeneity of triple negative breast cancer: Current advances in subtyping and treatment implications. J Exp Clin Cancer Res. 2022;41:265.
Palazzo AF, Koonin EV. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell. 2020;183:1151–61.
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22.
Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, et al. Translation of noncoding RNAs and cancer. Cancer Lett. 2021;497:89–99.
Pang Y, Liu Z, Han H, Wang B, Li W, Mao C, et al. Peptide SMIM30 promotes HCC development by inducing SRC/YES1 membrane anchoring and MAPK pathway activation. J Hepatol. 2020;73:1155–69.
Huang JZ, Chen M, Chen D, Gao XC, Zhu S, Huang H, et al. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell. 2017;68:171–184.e6.
Marciniak SJ, Chambers JE, Ron D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat Rev Drug Discov. 2022;21:115–40.
Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21:421–38.
Batista A, Rodvold JJ, Xian S, Searles SC, Lew A, Iwawaki T, et al. IRE1alpha regulates macrophage polarization, PD-L1 expression, and tumor survival. PLoS Biol. 2020;18:e3000687.
Shin GC, Moon SU, Kang HS, Choi HS, Han HD, Kim KH. PRKCSH contributes to tumorigenesis by selective boosting of IRE1 signaling pathway. Nat Commun. 2019;10:3185.
Glimcher LH, Lee AH, Iwakoshi NN. XBP-1 and the unfolded protein response (UPR). Nat Immunol. 2020;21:963–5.
Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–59.
Chen X, Iliopoulos D, Zhang Q, Tang Q, Greenblatt MB, Hatziapostolou M, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–7.
McCarthy N. Breast cancer: Hypoxia and XBP1S. Nat Rev Cancer. 2014;14:295.
Li S, Chen X, Liu X, Yu Y, Pan H, Haak R, et al. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9.
Wang X, Yin H, Zhang L, Zheng D, Yang Y, Zhang J, et al. The construction and analysis of the aberrant lncRNA-miRNA-mRNA network in non-small cell lung cancer. J Thorac Dis. 2019;11:1772–8.
Gong A, Luo X, Tan Y, Chen H, Luo G. High expression of C10orf91 and LINC01224 in hepatocellular carcinoma and poor prognosis. Am J Transl Res. 2022;14:2567–79.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu WP, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24.
Zheng J, Wang Q, Chen J, Cai G, Zhang Z, Zou H, et al. Tumor mitochondrial oxidative phosphorylation stimulated by the nuclear receptor RORgamma represents an effective therapeutic opportunity in osteosarcoma. Cell Rep Med. 2024;5:101519.
Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: the new central dogma of cancer biology. Sci China Life Sci. 2021;64:22–50.
Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–57.
Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045.
Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509.
Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965–81.
Mas AM, Huarte M. Long Noncoding RNA Signatures as Cancer Biomarkers. Journal of Clinical Oncology. 2023;41:3059.
Mercer TR, Munro T, Mattick JS. The potential of long noncoding RNA therapies. Trends Pharmacol Sci. 2022;43:269–80.
Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606.
Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–32.
Ge Q, Jia D, Cen D, Qi Y, Shi C, Li J, et al. Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J Clin Investig. 2021;131:e152911.
Guo M, Zhuang H, Huang J, Shao X, Bai N, Li M, et al. LINC02870 facilitates SNAIL translation to promote hepatocellular carcinoma progression. Mol Cell Biochem. 2023;478:1899–914.
Urra H, Dufey E, Avril T, Chevet E, Hetz C. Endoplasmic Reticulum Stress and the Hallmarks of Cancer. Trends Cancer. 2016;2:252–62.
Chen X, Cubillos-Ruiz JR. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat Rev Cancer. 2021;21:71–88.
Chen W, Wu C, Chen Y, Guo Y, Qiu L, Liu Z, et al. Downregulation of ceramide synthase 1 promotes oral cancer through endoplasmic reticulum stress. Int J Oral Sci. 2021;13:10.
Zhang Y, Liu Y, Zhou Y, Zheng Z, Tang W, Song M, et al. Lentinan inhibited colon cancer growth by inducing endoplasmic reticulum stress-mediated autophagic cell death and apoptosis. Carbohydr Polym. 2021;267:118154.
Chen X, Shi C, He M, Xiong S, Xia X. Endoplasmic reticulum stress: molecular mechanism and therapeutic targets. Signal Transduct Target Ther. 2023;8:352.
Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–61.
Valdes P, Mercado G, Vidal RL, Molina C, Parsons G, Court FA, et al. Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proc Natl Acad Sci USA. 2014;111:6804–9.
Grandjean JMD, Madhavan A, Cech L, Seguinot BO, Paxman RJ, Smith E, et al. Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nat Chem Biol. 2020;16:1052–61.
Madhavan A, Kok BP, Rius B, Grandjean JMD, Alabi A, Albert V, et al. Pharmacologic IRE1/XBP1s activation promotes systemic adaptive remodeling in obesity. Nat Commun. 2022;13:608.
Lhomond S, Avril T, Dejeans N, Voutetakis K, Doultsinos D, McMahon M, et al. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol Med. 2018;10:e7929.
Sheng X, Nenseth HZ, Qu S, Kuzu OF, Frahnow T, Simon L, et al. IRE1alpha-XBP1s pathway promotes prostate cancer by activating c-MYC signaling. Nat Commun. 2019;10:323.
Zhao Y, Zhang W, Huo M, Wang P, Liu X, Wang Y, et al. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduct Target Ther. 2021;6:357.
Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–81.
Raymundo DP, Doultsinos D, Guillory X, Carlesso A, Eriksson LA, Chevet E. Pharmacological Targeting of IRE1 in Cancer. Trends Cancer. 2020;6:1018–30.
Rajapaksa G, Nikolos F, Bado I, Clarke R, Gustafsson JA, Thomas C. ERbeta decreases breast cancer cell survival by regulating the IRE1/XBP-1 pathway. Oncogene. 2015;34:4130–41.
Zhao N, Cao J, Xu L, Tang Q, Dobrolecki LE, Lv X, et al. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J Clin Investig. 2018;128:1283–99.
Barua D, Gupta A, Gupta S. Targeting the IRE1-XBP1 axis to overcome endocrine resistance in breast cancer: Opportunities and challenges. Cancer Lett. 2020;486:29–37.
Cairrao F, Santos CC, Le Thomas A, Marsters S, Ashkenazi A, Domingos PM. Pumilio protects Xbp1 mRNA from regulated Ire1-dependent decay. Nat Commun. 2022;13:1587.
El Manaa W, Duplan E, Goiran T, Lauritzen I, Vaillant Beuchot L, Lacas-Gervais S, et al. Transcription- and phosphorylation-dependent control of a functional interplay between XBP1s and PINK1 governs mitophagy and potentially impacts Parkinson disease pathophysiology. Autophagy. 2021;17:4363–85.
Acknowledgements
We thank all members of JJW’s lab for helpful discussions and input. This research was supported by the National Natural Science Foundation of China (82273956, 82272968); the Guangdong Basic and Applied Basic Research Foundation (2022B1515130008); the Key Research and Development Plan of Guangzhou City (202206080007); and the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (No. 23ptpy36, 24xkjc016).
Author information
Authors and Affiliations
Contributions
XW: conducted the protein-identification experiments, prepared the figures and wrote the original manuscript. QW: conducted the functional experiments, and prepared the figures. HW: conducted the lentiviral packaging and animal experiments. GC and YA: participated in protein expression and purification. PL and HZ: provided funding acquisition and project administration. HC: provided materials and supervision. SJ: provided funding acquisition. JY: wrote and revised the manuscript. JW: designed the experiments, funding acquisition, and supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Animal experimental procedures and operations were approved by the Animal Welfare and Ethics Committee of Sun Yat-sen University, complied with the ARRIVE guidelines, and carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Wang, Q., Wang, H. et al. Small protein ERSP encoded by LINC02870 promotes triple negative breast cancer progression via IRE1α/XBP1s activation. Cell Death Differ 32, 1014–1025 (2025). https://doi.org/10.1038/s41418-025-01443-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01443-5
This article is cited by
-
Recent advances in structural and functional diversities of cancer lncRNA-encoded peptides: current opportunities and challenges for enhancing cancer diagnosis and treatment
Cancer Cell International (2026)
-
KIRA6 restrains the generation of myeloid-derived suppressor cells and overcomes resistance to anti-PD-1 therapy
Cell Death & Disease (2025)