Abstract
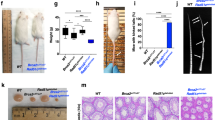
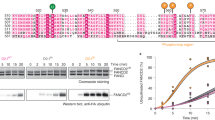
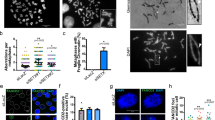
Fanconi Anemia (FA) is an autosomal recessive disorder characterized by diverse clinical manifestations such as aplastic anemia, cancer predisposition, and developmental defects including hypogonadism, microcephaly, organ dysfunction, infertility, hyperpigmentation, microphthalmia, and skeletal defects. In addition to the well-described defects in DNA repair, mitochondrial dysfunction due to defects in mitochondrial autophagy (mitophagy) is also associated with FA, although its contribution to FA phenotypes is unknown. This study focused on the FANCC gene, which, alongside other FA genes, is integral to DNA repair and mitochondrial quality control. In the present study, we created a FANCC mutant mouse model, based on a human mutation (FANCC c.67delG) that is defective in DNA repair but proficient in mitophagy. We found that the FANCC c.67delG mutant mouse model recapitulates some phenotypes observed in FA patients, such as cellular hypersensitivity to DNA cross-linking agents and hematopoietic defects. In contrast, FA phenotypes such as microphthalmia, hypogonadism, and infertility, present in FANCC-deficient mice, were absent in the FANCC c.67delG mice, suggesting that the N-terminal 55 amino acids of FANCC are dispensable for these developmental processes. Furthermore, the FANCC c.67delG mutation preserved mitophagy, and unlike the FANCC null mutation, did not lead to the accumulation of damaged mitochondria in cells or tissues. This study highlights the multifaceted nature of the FANCC protein, with distinct domains responsible for DNA repair and mitophagy. Our results suggest that developmental defects in FA may not solely stem from DNA repair deficiencies but could also involve other functions, such as mitochondrial quality control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10.
Alter BP. Diagnosis, genetics, and management of inherited bone marrow failure syndromes. Hematol Am Soc Hematol Educ Program. 2007:29–39.
Moldovan GL, D’Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49.
Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–408.
Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668:133–40.
Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet. 2009;18:3484–95.
Whitney MA, Royle G, Low MJ, Kelly MA, Axthelm MK, Reifsteck C, et al. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58.
Chen M, Tomkins DJ, Auerbach W, McKerlie C, Youssoufian H, Liu L, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12:448–51.
Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–22.
Niraj J, Färkkilä A. D’Andrea AD. The Fanconi anemia pathway in cancer. Annu Rev Cancer Biol. 2019;3:457–78.
Noll M, Battaile KP, Bateman R, Lax TP, Rathbun K, Reifsteck C, et al. Fanconi anemia group A and C double-mutant mice: functional evidence for a multi-protein Fanconi anemia complex. Exp Hematol. 2002;30:679–88.
Koomen M, Cheng NC, van de Vrugt HJ, Godthelp BC, van der Valk MA, Oostra AB, et al. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum Mol Genet. 2002;11:273–81.
Yang Y, Kuang Y, Montes De Oca R, Hays T, Moreau L, et al. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001;98:3435–40.
Cheng NC, van de Vrugt HJ, van der Valk MA, Oostra AB, Krimpenfort P, de Vries Y, et al. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet 2000;9:1805–11.
Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–35.
Pagano G, Tiano L, Pallardó FV, Lyakhovich A, Mukhopadhyay SS, Di Bartolomeo P, et al. Re-definition and supporting evidence toward Fanconi Anemia as a mitochondrial disease: prospects for new design in clinical management. Redox Biol. 2021;40:101860.
Savoia A, Garcia-Higuera I, D’Andrea AD. Nuclear localization of the Fanconi anemia protein FANCC is required for functional activity. Blood. 1999;93:4025–6.
Sumpter R Jr, Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, et al. Fanconi anemia proteins function in mitophagy and immunity. Cell. 2016;165:867–81.
Haneline LS, Broxmeyer HE, Cooper S, Hangoc G, Carreau M, Buchwald M, et al. Multiple inhibitory cytokines induce deregulated progenitor growth and apoptosis in hematopoietic cells from Fac-/- mice. Blood. 1998;91:4092–8.
Pagano G, Talamanca AA, Castello G, d’Ischia M, Pallardó FV, Petrović S, et al. From clinical description, to in vitro and animal studies, and backward to patients: oxidative stress and mitochondrial dysfunction in Fanconi anemia. Free Radic Biol Med. 2013;58:118–25.
Hoshino T, Wang J, Devetten MP, Iwata N, Kajigaya S, Wise RJ, et al. Molecular chaperone GRP94 binds to the Fanconi anemia group C protein and regulates its intracellular expression. Blood. 1998;91:4379–86.
Garbati MR, Hays LE, Keeble W, Yates JE, Rathbun RK, Bagby GC. FANCA and FANCC modulate TLR and p38 MAPK-dependent expression of IL-1β in macrophages. Blood. 2013;122:3197–205.
Whitney MA, Saito H, Jakobs PM, Gibson RA, Moses RE, Grompe M. A common mutation in the FACC gene causes Fanconi anaemia in Ashkenazi Jews. Nat Genet. 1993;4:202–5.
Yamashita T, Wu N, Kupfer G, Corless C, Joenje H, Grompe M. D’Andrea AD. Clinical variability of Fanconi anemia (type C) results from expression of an amino terminal truncated Fanconi anemia complementation group C polypeptide with partial activity. Blood. 1996;87:4424–32.
Faivre L, Guardiola P, Lewis C, Dokal I, Ebell W, Zatterale A, et al. Association of complementation group and mutation type with clinical outcome in Fanconi anemia. European Fanconi Anemia Research Group. Blood. 2000;96:4064–70.
Gillio AP, Verlander PC, Batish SD, Giampietro PF, Auerbach AD. Phenotypic consequences of mutations in the Fanconi anemia FAC gene: an International Fanconi Anemia Registry study. Blood. 1997;90:105–10.
García-de Teresa B, Frias S, Molina B, Villarreal MT, Rodriguez A, Carnevale A, et al. FANCC Dutch founder mutation in a Mennonite family from Tamaulipas, México. Mol Genet Genom Med. 2019;7:e710.
de Vries Y, Lwiwski N, Levitus M, Kuyt B, Israels SJ, Arwert F, et al. A Dutch Fanconi Anemia FANCC founder mutation in Canadian Manitoba Mennonites. Anemia. 2012;2012:865170.
Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DY, Tothill RW, et al. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8:e1002894.
Information. NNCfB. ClinVar; [VCV000012049.45], [National Center for Biotechnology Information. ClinVar; [VCV000012049.45], https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV.45 (accessed May 13, 2024)].
Pang Q, Christianson TA, Keeble W, Diaz J, Faulkner GR, Reifsteck C, et al. The Fanconi anemia complementation group C gene product: structural evidence of multifunctionality. Blood. 2001;98:1392–401.
Calado RT, Clé DV. Treatment of inherited bone marrow failure syndromes beyond transplantation. Hematol Am Soc Hematol Educ Program. 2017;2017:96–101.
Oppezzo A, Bourseguin J, Renaud E, Pawlikowska P, Rosselli F. Microphthalmia transcription factor expression contributes to bone marrow failure in Fanconi anemia. J Clin Invest. 2020;130:1377–91.
Sejas DP, Rani R, Qiu Y, Zhang X, Fagerlie SR, Nakano H, et al. Inflammatory reactive oxygen species-mediated hemopoietic suppression in Fancc-deficient mice. J Immunol. 2007;178:5277–87.
Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14.
Bagby GC. Multifunctional Fanconi proteins, inflammation and the Fanconi phenotype. EBioMedicine. 2016;8:10–1.
Shyamsunder P, Esner M, Barvalia M, Wu YJ, Loja T, Boon HB, et al. Impaired mitophagy in Fanconi anemia is dependent on mitochondrial fission. Oncotarget. 2016;7:58065–74.
Solanki A, Rajendran A, Mohan S, Raj R, Vundinti BR. Mitochondrial DNA variations and mitochondrial dysfunction in Fanconi anemia. PLoS One. 2020;15:e0227603.
Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, et al. Measuring in vivo mitophagy. Mol Cell. 2015;60:685–96.
Freie B, Li X, Ciccone SL, Nawa K, Cooper S, Vogelweid C, et al. Fanconi anemia type C and p53 cooperate in apoptosis and tumorigenesis. Blood. 2003;102:4146–52.
Rani R, Li J, Pang Q. Differential p53 engagement in response to oxidative and oncogenic stresses in Fanconi anemia mice. Cancer Res. 2008;68:9693–702.
Skeie JM, Nishimura DY, Wang CL, Schmidt GA, Aldrich BT, Greiner MA. Mitophagy: an emerging target in ocular pathology. Investig Ophthalmol Vis Sci. 2021;62:22.
Fraser JA, Biousse V, Newman NJ. The neuro-ophthalmology of mitochondrial disease. Surv Ophthalmol. 2010;55:299–334.
Agerholm JS, DeLay J, Hicks B, Fredholm M. First confirmed case of the bovine brachyspina syndrome in Canada. Can Vet J. 2010;51:1349–50.
Peake JD, Noguchi C, Lin B, Theriault A, O’Connor M, Sheth S, et al. FANCD2 limits acetaldehyde-induced genomic instability during DNA replication in esophageal keratinocytes. Mol Oncol. 2021;15:3109–24.
Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–5.
Acknowledgements
We would like to thank the Transgenic and Gene Knockout Shared Resource at St. Jude Children’s Research Hospital, Memphis, TN, for technical assistance.
Funding
This work was supported by NCI R35 231620 (D.R.G.) and the John H Sununu named Fellowship (S.B).
Author information
Authors and Affiliations
Contributions
S.B. contributed to conceptualization, performed experiments and data analysis, writing, review, and editing of the manuscript. C.G. performed experiments and analysis of imaging results, S.S. performed experiments, M.Y provided intellectual input, R.S. contributed to conceptualization, project administration, resource provision, and supervision. H.S analyzed results including pathology evaluations and provided technical support, S.P contributed to the methodology for the mutant mice generation, M.W provided Intellectual input and reviewed the manuscript, D.R.G contributed to conceptualization, funding, project administration, resources, supervision, writing, review, and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
During the course of this research, D.R.G. consulted for or received support from Amgen, Ventus, ASHA, Boehringer Ingelheim, Mirumus, and Sonata. The authors declare no conflicts of interest.
Ethics approval and consent to participate
All Mouse studies were conducted in accordance with protocols approved by the St. Jude Children’s Research Hospital Committee on Care and Use of Animals and in compliance with all relevant ethical guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Beesetti, S., Guy, C., Sirasanagandla, S. et al. Distinct developmental outcomes in DNA repair-deficient FANCC c.67delG mutant and FANCC−/− Mice. Cell Death Differ 32, 1294–1302 (2025). https://doi.org/10.1038/s41418-025-01461-3
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01461-3