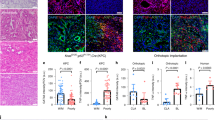
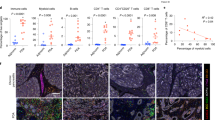
Abstract
Alternative cleavage and polyadenylation (APA) have gained increasing attention in cancer biology, yet its role in modulating anti-tumor immune response remains largely unexplored. Here, we identify the cleavage stimulation factor 2 (CSTF2), an APA-related gene, as a pivotal suppressor of anti-tumor immunity in pancreatic ductal adenocarcinoma (PDAC). CSTF2 promotes tumor development by inhibiting the infiltration and cytotoxic immune cell recruitment function of TCRαβ+CD4−CD8−NK1.1− innate αβ T (iαβT) cells. Mechanistically, CSTF2 diminishes CXCL10 expression by promoting PolyA polymerase alpha (PAPα) binding to the 3’ untranslated regions of CXCL10 RNA, resulting in shortened PolyA tails and compromised RNA stability. Furthermore, we identify Forsythoside B, a selective inhibitor targeting the RNA recognition motif of CSTF2, can effectively activate anti-tumor immunity and overcome resistance to immune checkpoint blockade (ICB) therapy. Collectively, our findings unveil CSTF2 as a promising therapeutic target for sensitizing PDAC to ICB therapy.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The bulk RNA sequence data from 65 PDAC patients have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences http://bigd.big.ac.cn/ under restricted access: HRA000095. The data of Single-cell RNA-seq in this study have been deposited at Genome Sequence Archive (GSA) with accession numbers: GSE271287. The original contributions presented in the study are included in the article or the supplementary material.
References
Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20:599–614.
Xiong M, Chen L, Zhou L, Ding Y, Kazobinka G, Chen Z, et al. NUDT21 inhibits bladder cancer progression through ANXA2 and LIMK2 by alternative polyadenylation. Theranostics. 2019;9:7156–67.
Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–6.
Wu Z, Jia X, de la Cruz L, Su XC, Marzolf B, Troisch P, et al. Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity. 2008;29:863–75.
Lin W, Kurosawa K, Murayama A, Kagaya E, Ohta K. B-cell display-based one-step method to generate chimeric human IgG monoclonal antibodies. Nucleic Acids Res. 2011;39:e14.
Chen X, Zhang JX, Luo JH, Wu S, Yuan GJ, Ma NF, et al. CSTF2-induced shortening of the RAC1 3’UTR promotes the pathogenesis of urothelial carcinoma of the bladder. Cancer Res. 2018;78:5848–62.
Xu Y, Yuan F, Sun Q, Zhao L, Hong Y, Tong S, et al. The RNA-binding protein CSTF2 regulates BAD to inhibit apoptosis in glioblastoma. Int J Biol Macromol. 2023;226:915–26.
Akman HB, Oyken M, Tuncer T, Can T, Erson-Bensan AE. 3’UTR shortening and EGF signaling: implications for breast cancer. Hum Mol Genet. 2015;24:6910–20.
Adamska A, Domenichini A, Falasca M. Pancreatic ductal adenocarcinoma: Current and evolving therapies. Int J Mol Sci. 2017;18:1338.
Liu C, Yang M, Zhang D, Chen M, Zhu D. Clinical cancer immunotherapy: Current progress and prospects. Front Immunol. 2022;13:961805.
Zhu YH, Zheng JH, Jia QY, Duan ZH, Yao HF, Yang J, et al. Immunosuppression, immune escape, and immunotherapy in pancreatic cancer: focused on the tumor microenvironment. Cell Oncol (Dordr). 2023;46:17–48.
Wu Z, Zheng Y, Sheng J, Han Y, Yang Y, Pan H, et al. CD3(+)CD4(-)CD8(-) (Double-Negative) T Cells in inflammation, immune disorders and cancer. Front Immunol. 2022;13:816005.
Chen J, Hu P, Wu G, Zhou H. Antipancreatic cancer effect of DNT cells and the underlying mechanism. Pancreatology. 2019;19:105–13.
Hundeyin M, Kurz E, Mishra A, Rossi JAK, Liudahl SM, Leis KR, et al. Innate alphabeta T cells mediate antitumor immunity by orchestrating immunogenic macrophage programming. Cancer Discov. 2019;9:1288–305.
Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49:178–93.e7.
Yao J, Ly D, Dervovic D, Fang L, Lee JB, Kang H, et al. Human double negative T cells target lung cancer via ligand-dependent mechanisms that can be enhanced by IL-15. J Immunother Cancer. 2019;7:17.
Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4.
Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol. 2018;19:1112–25.
Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell. 2019;35:559–72.e7.
Lv J, Wei Y, Yin JH, Chen YP, Zhou GQ, Wei C, et al. The tumor immune microenvironment of nasopharyngeal carcinoma after gemcitabine plus cisplatin treatment. Nat Med. 2023;29:1424–36.
Ren X, Yang X, Cheng B, Chen X, Zhang T, He Q, et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun. 2017;8:14053.
Daley D, Zambirinis CP, Seifert L, Akkad N, Mohan N, Werba G, et al. gammadelta T Cells support pancreatic oncogenesis by restraining alphabeta T Cell activation. Cell. 2016;166:1485–99.e15.
Eil R, Vodnala SK, Clever D, Klebanoff CA, Sukumar M, Pan JH, et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature. 2016;537:539–43.
Oveland E, Karlsen TV, Haslene-Hox H, Semaeva E, Janaczyk B, Tenstad O, et al. Proteomic evaluation of inflammatory proteins in rat spleen interstitial fluid and lymph during LPS-induced systemic inflammation reveals increased levels of ADAMST1. J Proteome Res. 2012;11:5338–49.
Wang Y, Luu LDW, Liu S, Zhu X, Huang S, Li F, et al. Single-cell transcriptomic analysis reveals a systemic immune dysregulation in COVID-19-associated pediatric encephalopathy. Signal Transduct Target Ther. 2023;8:398.
Wang Y, Chen H, Liu W, Yan H, Zhang Y, Cheung AHK, et al. MCM6 is a critical transcriptional target of YAP to promote gastric tumorigenesis and serves as a therapeutic target. Theranostics. 2022;12:6509–26.
Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–7.
Shao M, Lu T, Zhang C, Zhang YZ, Kong SH, Shi DL. Rbm24 controls poly(A) tail length and translation efficiency of crystallin mRNAs in the lens via cytoplasmic polyadenylation. Proc Natl Acad Sci USA. 2020;117:7245–54.
Chen Z, Luo Z, Zhang D, Li H, Liu X, Zhu K, et al. TIGER: A web portal of tumor immunotherapy gene expression resource. Genomics Proteomics Bioinformatics. 2023;21:337–48.
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612.
Koferle A, Schlattl A, Hormann A, Thatikonda V, Popa A, Spreitzer F, et al. Interrogation of cancer gene dependencies reveals paralog interactions of autosome and sex chromosome-encoded genes. Cell Rep. 2022;39:110636.
Liu J, Guan X, Ma X. Interferon regulatory factor 1 is an essential and direct transcriptional activator for interferon γ-induced RANTES/CCl5 expression in macrophages. J Biol Chem. 2005;280:24347–55.
Zeng Z, Veitch M, Kelly GA, Tuong ZK, Cruz JG, Frazer IH, et al. IFN-γ critically enables the intratumoural infiltration of CXCR3(+) CD8(+) T Cells to drive squamous cell carcinoma regression. Cancers (Basel) 2021;13:2131.
Gocher AM, Workman CJ, Vignali DAA. Interferon-γ: teammate or opponent in the tumour microenvironment? Nat Rev Immunol. 2022;22:158–72.
Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, et al. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci USA. 2012;109:18773–8.
Laishram RS. Poly(A) polymerase (PAP) diversity in gene expression-star-PAP vs canonical PAP. FEBS Lett. 2014;588:2185–97.
Li W, Laishram RS, Ji Z, Barlow CA, Tian B, Anderson RA. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIα and PKCδ signaling. Mol Cell. 2012;45:25–37.
Kandala DT, Mohan N, A V, A PS, G R, Laishram RS. CstF-64 and 3’-UTR cis-element determine Star-PAP specificity for target mRNA selection by excluding PAPα. Nucleic Acids Res. 2016;44:811–23.
Passmore LA, Coller J. Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol. 2022;23:93–106.
Kamath SD, Kalyan A, Kircher S, Nimeiri H, Fought AJ, Benson A, et al. Ipilimumab and gemcitabine for advanced pancreatic cancer: A Phase Ib study. Oncologist. 2020;25:e808–e15.
O’Reilly EM, Oh DY, Dhani N, Renouf DJ, Lee MA, Sun W, et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: A phase 2 randomized clinical trial. JAMA Oncol. 2019;5:1431–8.
Muller M, Haghnejad V, Schaefer M, Gauchotte G, Caron B, Peyrin-Biroulet L, et al. The immune landscape of human pancreatic ductal carcinoma: Key players, clinical implications, and challenges. Cancers (Basel) 2022;14:995.
Ullman NA, Burchard PR, Dunne RF, Linehan DC. Immunologic strategies in pancreatic cancer: Making cold tumors hot. J Clin Oncol. 2022;40:2789–805.
Schizas D, Charalampakis N, Kole C, Economopoulou P, Koustas E, Gkotsis E, et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev. 2020;86:102016.
Bear AS, Vonderheide RH. O’Hara MH. Challenges and opportunities for pancreatic cancer immunotherapy. Cancer Cell. 2020;38:788–802.
Yeh HS, Yong J. Alternative polyadenylation of mRNAs: 3’-untranslated region matters in gene expression. Mol Cells. 2016;39:281–5.
Li L, Ma X, Cui Y, Rotival M, Chen W, Zou X, et al. Immune-response 3’UTR alternative polyadenylation quantitative trait loci contribute to variation in human complex traits and diseases. Nat Commun. 2023;14:8347.
Romero JM, Grünwald B, Jang GH, Bavi PP, Jhaveri A, Masoomian M, et al. A four-chemokine signature is associated with a T-cell-inflamed phenotype in primary and metastatic pancreatic cancer. Clin Cancer Res. 2020;26:1997–2010.
Yang MW, Tao LY, Jiang YS, Yang JY, Huo YM, Liu DJ, et al. Perineural invasion reprograms the immune microenvironment through cholinergic signaling in pancreatic ductal adenocarcinoma. Cancer Res. 2020;80:1991–2003.
Zhang W, Wan Y, Zhang Y, Liu Q, Zhu X. CSTF2 acts as a prognostic marker correlated with immune infiltration in hepatocellular carcinoma. Cancer Manag Res. 2022;14:2691–709.
Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat Rev Clin Oncol. 2020;17:527–40.
Zheng Y, Li X, Deng S, Zhao H, Ye Y, Zhang S, et al. CSTF2 mediated mRNA N6-methyladenosine modification drives pancreatic ductal adenocarcinoma m6A subtypes. Nat Commun. 2023;14:6334.
Ramakrishnan P, Loh WM, Gopinath SCB, Bonam SR, Fareez IM, Mac Guad R, et al. Selective phytochemicals targeting pancreatic stellate cells as new anti-fibrotic agents for chronic pancreatitis and pancreatic cancer. Acta Pharm Sin B. 2020;10:399–413.
Ma X, Chan TA. Solving the puzzle of what makes immunotherapies work. Trends Cancer. 2022;8:890–900.
Acknowledgements
The authors would like to acknowledge Prof. Dongming Kuang (college of life science in Sun Yat-sen University) for his insightful advice during the project.
Funding
This study was supported by National Natural Science Foundation of China Projects (82325037 and 82072617 to J Zheng, 82272694 to X Huang, 82003162 to J Zhang, and 82303159 to CX), the National Key R&D Program of China (2021YFA1302100 to J Zheng), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096 to DL) and Cancer Innovative Research Programme of Sun Yat-sen University Cancer Center (CIRP-SYSUCC-0002 and PT13010201 to DL), Young Talents Programme of Sun Yat-sen University Cancer Center (YTP-SYSUCC-0062 to J Zhang, YTP-SYSUCC0015 to J Zheng and YTP-SYSUCC-0068 to X Huang).
Author information
Authors and Affiliations
Contributions
X He, JL, J Zhang, J Zheng, DL and X Huang designed the study. X He, SZ and JL contributed to the performance of experiments, X He and YZ contributed to the interpretation of data. X He, J Zhang, J Zheng and DL contributed to the writing of the paper. ZC, ZX, CX, L Zeng, Shuang Liu, Shaoqiu Liu, RB and SW contributed to the methods and preparation of figure. L Zhuang, ML, HZ and QZ were responsible for tissue samples and matched clinical information collection. J Zhang, X Huang, J Zheng and DL supervised the work. For the co–first authors, the authorship order was assigned on the basis of their contributions to this work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All methods in this study were performed in accordance with the relevant guidelines and regulations. Informed consent was obtained from each patient, and this study was approved by the Institutional Review Board of the Sun Yat-sen University Cancer Center (approval number: GZR2020-036). All animal experiments were approved by the Institutional Animal Care and Use Committee of the Sun Yat-sen University Cancer Center (approval number: 2021-000284).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, X., Liu, J., Zhou, Y. et al. CSTF2-impeded innate αβ T cell infiltration and activation exacerbate immune evasion of pancreatic cancer. Cell Death Differ 32, 973–988 (2025). https://doi.org/10.1038/s41418-025-01464-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01464-0