Abstract
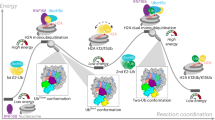
The ubiquitination of histone H2A/H2AX, catalyzed by RNF8/RNF168, is a crucial step in the repair of DNA double-strand breaks (DSBs), playing a significant role in transmitting and amplifying DNA damage response signals. However, the upstream regulatory mechanisms of RNF168 remain unclear. Here, we demonstrate that ZNF451 catalyzes the SUMOylation of RNF168, thereby regulating the ubiquitination of histone H2A/H2AX. Specifically, ZNF451 rapidly responds to radiation-induced DNA damage, accumulating abundantly at damage sites and catalyzing the SUMO2 modification of RNF168. This modification stabilizes RNF168, enhancing its accumulation at damage sites, which increases the ubiquitination levels of downstream histone H2A/H2AX and promotes the DNA damage repair process. Furthermore, we find that ZNF451 and RNF8 jointly regulate RNF168 in a novel manner, exhibiting both competitive and cooperative characteristics. The interaction between RNF168 and either ZNF451 or RNF8 mutually inhibits each other. However, simultaneous loss of ZNF451 and RNF8 markedly impedes the recruitment of RNF168 to damage sites. Whereas, varying expression levels of ZNF451 and RNF8 suggest that both facilitate the interaction between RNF168 and the downstream factor H2AX, but the interaction plateaus beyond a specific threshold. Altogether, these findings reveal that the SUMOylation catalyzed by ZNF451 is involved in regulating RNF168-induced ubiquitin signaling in DSBs repair and suggest that ZNF451 could serve as a potential therapeutic target in tumor radiotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Other data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Tang L, Wei F, Wu Y, He Y, Shi L, Xiong F, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87.
An L, Li M, Jia Q. Mechanisms of radiotherapy resistance and radiosensitization strategies for esophageal squamous cell carcinoma. Mol Cancer. 2023;22:140.
Wu Y, Song Y, Wang R, Wang T. Molecular mechanisms of tumor resistance to radiotherapy. Mol Cancer. 2023;22:96.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8.
White RR, Vijg J. Do DNA Double-Strand Breaks Drive Aging? Mol Cell. 2016;63:729–38.
Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–54.
Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–95.
Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14.
Krais JJ, Wang Y, Patel P, Basu J, Bernhardy AJ, Johnson N. RNF168-mediated localization of BARD1 recruits the BRCA1-PALB2 complex to DNA damage. Nat Commun. 2021;12:5016.
Becker JR, Clifford G, Bonnet C, Groth A, Wilson MD, Chapman JR. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature. 2021;596:433–7.
Zhang T, Yang H, Zhou Z, Bai Y, Wang J, Wang W. Crosstalk between SUMOylation and ubiquitylation controls DNA end resection by maintaining MRE11 homeostasis on chromatin. Nat Commun. 2022;13:5133.
Hariharasudhan G, Jeong SY, Kim MJ, Jung SM, Seo G, Moon JR, et al. TOPORS-mediated RAD51 SUMOylation facilitates homologous recombination repair. Nucleic Acids Res. 2022;50:1501–16.
Zhu S, Hou J, Gao H, Hu Q, Kloeber JA, Huang J, et al. SUMOylation of HNRNPA2B1 modulates RPA dynamics during unperturbed replication and genotoxic stress responses. Mol Cell. 2023;83:539–55 e7.
Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90.
Luo K, Zhang H, Wang L, Yuan J, Lou Z. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 2012;31:3008–19.
Danielsen JR, Povlsen LK, Villumsen BH, Streicher W, Nilsson J, Wikstrom M, et al. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J Cell Biol. 2012;197:179–87.
Vertegaal ACO. Signalling mechanisms and cellular functions of SUMO. Nat Rev Mol Cell Biol. 2022;23:715–31.
Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–93.
Zeng W, Gu S, Yu Y, Feng Y, Xiao M, Feng XH. ZNF451 stabilizes TWIST2 through SUMOylation and promotes epithelial-mesenchymal transition. Am J Cancer Res. 2021;11:898–915.
Feng Y, Wu H, Xu Y, Zhang Z, Liu T, Lin X, et al. Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-beta signaling. J Biol Chem. 2014;289:2072–83.
Karvonen U, Jaaskelainen T, Rytinki M, Kaikkonen S, Palvimo JJ. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J Mol Biol. 2008;382:585–600.
Schellenberg MJ, Lieberman JA, Herrero-Ruiz A, Butler LR, Williams JG, Munoz-Cabello AM, et al. ZATT (ZNF451)-mediated resolution of topoisomerase 2 DNA-protein cross-links. Science. 2017;357:1412–6.
Tian T, Bu M, Chen X, Ding L, Yang Y, Han J, et al. The ZATT-TOP2A-PICH Axis Drives Extensive Replication Fork Reversal to Promote Genome Stability. Mol Cell. 2021;81:198–211 e6.
Zagnoli-Vieira G, Caldecott KW. TDP2, TOP2, and SUMO: what is ZATT about? Cell Res. 2017;27:1405–6.
Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714.
Xie X, Hu H, Tong X, Li L, Liu X, Chen M, et al. The mTOR-S6K pathway links growth signalling to DNA damage response by targeting RNF168. Nat Cell Biol. 2018;20:320–31.
Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709.
Sharma A, Alswillah T, Singh K, Chatterjee P, Willard B, Venere M, et al. USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy. 2018;14:1976–90.
Bai Y, Wang W, Li S, Zhan J, Li H, Zhao M, et al. C1QBP promotes homologous recombination by stabilizing MRE11 and controlling the assembly and activation of MRE11/RAD50/NBS1 complex. Mol Cell. 2019;75:1299–314 e6.
Wang J, Xu WH, Wei Y, Zhu Y, Qin XJ, Zhang HL, et al. Elevated MRE11 expression associated with progression and poor outcome in prostate cancer. J Cancer. 2019;10:4333–40.
Ho V, Chung L, Wilkinson K, Lea V, Lim SH, Abubakar A, et al. Prognostic significance of MRE11 overexpression in colorectal cancer patients. Cancers (Basel). 2023;15:2438.
Alblihy A, Ali R, Algethami M, Shoqafi A, Toss MS, Brownlie J, et al. Targeting Mre11 overcomes platinum resistance and induces synthetic lethality in XRCC1 deficient epithelial ovarian cancers. NPJ Precis Oncol. 2022;6:51.
Liu Z, Zhang J, Xu J, Yang H, Li X, Hou Y, et al. RNF168 facilitates oestrogen receptor a transcription and drives breast cancer proliferation. J Cell Mol Med. 2018;22:4161–70.
Yu N, Xue M, Wang W, Xia D, Li Y, Zhou X, et al. RNF168 facilitates proliferation and invasion of esophageal carcinoma, possibly via stabilizing STAT1. J Cell Mol Med. 2019;23:1553–61.
Wang YY, Hung AC, Lo S, Hsieh YC, Yuan SF. MRE11 as a molecular signature and therapeutic target for cancer treatment with radiotherapy. Cancer Lett. 2021;514:1–11.
Xie T, Qin H, Yuan Z, Zhang Y, Li X, Zheng L. Emerging roles of RNF168 in tumor progression. Molecules. 2023;28:1417.
Zhang Y, Wang W, Min J, Liu S, Wang Q, Wang Y, et al. ZNF451 favors triple-negative breast cancer progression by enhancing SLUG-mediated CCL5 transcriptional expression. Cell Rep. 2023;42:112654.
Hodge CD, Ismail IH, Edwards RA, Hura GL, Xiao AT, Tainer JA, et al. RNF8 E3 ubiquitin ligase stimulates Ubc13 E2 conjugating activity that is essential for DNA double strand break signaling and BRCA1 tumor suppressor recruitment. J Biol Chem. 2016;291:9396–410.
Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–34.
Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Bottinger L, Klay D, et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300.
Hu Y, Sui X, Song F, Li Y, Li K, Chen Z, et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat Commun. 2021;12:2581.
Chen B, Xu F, Gao Y, Hu G, Zhu K, Lu H, et al. DNA damage-induced translocation of mitochondrial factor HIGD1A into the nucleus regulates homologous recombination and radio/chemo-sensitivity. Oncogene. 2022;41:1918–30.
Funding
This work was supported by the National Science Fund for Excellent Young Scholars (12122510), the National Natural Science Foundation of China (32171240, 12435019, 32401029 and 32271283), the HFIPS Director’s Fund (BJPY2021B07 and BJPY2023A010).
Author information
Authors and Affiliations
Contributions
FX, QX, Bin C and GZ designed and conceived of the study. FX, QX, Bin C, RW, JZ, XZ, ZZ, and Biao C conducted experiments. FX, ZY, JZ, SZ, XL, AX and LW analyzed and interpreted the data. FX and GZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Ethics statement
The study is approved by the Ethics Committee of Hefei Institutes of Physical Science, Chinese Academy of Sciences. All methods were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, F., Xia, Q., Chen, B. et al. ZNF451 collaborates with RNF8 to regulate RNF168 localization and amplify ubiquitination signaling to promote DNA damage repair and regulate radiosensitivity. Cell Death Differ 32, 1303–1316 (2025). https://doi.org/10.1038/s41418-025-01472-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01472-0