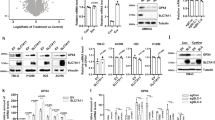
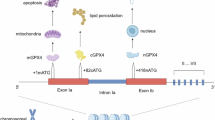
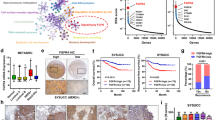
Abstract
Emerging evidence indicates that activation of ferroptosis by inhibition of glutathione peroxidase 4 (GPX4) may be exploited as a therapeutic strategy to suppress tumor growth and progression. However, application of GPX4 inhibitors in cancer treatment is hampered by their poor selectivity, which results in unfavorable toxicity. Herein, we identified GPX4 as a candidate for the autophagy pathway. We showed that GPX4 is ubiquitinated by TNF receptor-associated factor 6 (TRAF6), which promotes its recognition by p62 and leads to its selective autophagic degradation. Utilizing targeted protein degradation (TPD) approach, we developed a GPX4-targeted AUTAC and demonstrated that GPX4-AUTAC promoted the ubiquitination of GPX4, and enhanced the binding with GPX4 and p62, leading to the selective autophagy-dependent degradation of GPX4. Furthermore, GPX4-AUTAC treatment strongly induced ferroptosis and exhibited potent anti-cancer activity against breast cancer in vitro, in vivo, and patient-derived organoids (PDOs). Combination treatment of GPX4-AUTAC with sulfasalazine, a ferroptotic inducer, or chemotherapy drugs showed a synergistic anti-cancer effect against breast cancer. These results uncover a new targeted degradation strategy for GPX4 by inducing selective autophagy and provide a rationale for the use of GPX4-AUTAC as a novel therapeutic approach to treatment of breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Further inquiries can be directed to the corresponding author.
References
Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CACancer J Clin. 2024;74:12–49.
Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast. 2022;66:15–23.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Stockwell BR. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell. 2022;185:2401–21.
Lei G, Zhuang L, Gan BY. Targeting ferroptosis as a vulnerability in cancer. Nat Rev Cancer. 2022;22:381–96.
Bi G, Liang J, Bian Y, Shan G, Huang Y, Lu T, et al. Polyamine-mediated ferroptosis amplification acts as a targetable vulnerability in cancer. Nat Commun. 2024;15:2461.
Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31.
Ingold I, Berndt C, Schmitt S, Doll S, Poschmann G, Buday K, et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2018;172:409 .
Angeli JPF, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–U1120.
Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–7.
Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–50.
Chamberlain PP, Hamann LG. Development of targeted protein degradation therapeutics. Nat Chem Biol. 2019;15:937–44.
Dale B, Cheng M, Park KS, Kaniskan HÜ, Xiong Y, Jin J. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer. 2021;21:638–54.
Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42.
Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–85.
Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–42.
Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, Sato A, et al. AUTACs: cargo-specific degraders using selective autophagy. Mol Cell. 2019;76:797–810 e710.
Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–9.
Hou W, Xie YC, Song XX, Sun XF, Lotze MT, Zeh HJ, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–8.
Zhang ZL, Yao Z, Wang L, Ding H, Shao JJ, Chen AP, et al. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14:2083–103.
Yang M, Chen P, Liu J, Zhu S, Kroemer G, Klionsky DJ, et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv. 2019;5:eaaw2238.
Chen Y, Li Q, Li Q, Xing S, Liu Y, Liu Y, et al. p62/SQSTM1, a central but unexploited target: advances in its physiological/pathogenic functions and small molecular modulators. J Med Chem. 2020;63:10135–57.
Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2020;432:80–103.
Dósa A, Csizmadia T. The role of K63-linked polyubiquitin in several types of autophagy. Biol Futur. 2022;73:137–48.
Jiang XJ, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Bio. 2021;22:266–82.
Lei G, Zhuang L, Gan B. The roles of ferroptosis in cancer: tumor suppression, tumor microenvironment, and therapeutic interventions. Cancer Cell. 2024;42:513–34.
Vargas JNS, Hamasaki M, Kawabata T, Youle RJ, Yoshimori T. The mechanisms and roles of selective autophagy in mammals. Nat Rev Mol Cell Bio. 2022;24:167–85.
Lamark T, Johansen T. Mechanisms of selective autophagy. Annu Rev Cell Dev Biol. 2021;37:143–69.
Liu J, Kuang F, Kroemer G, Klionsky DJ, Kang R, Tang D. Autophagy-dependent ferroptosis: machinery and regulation. Cell Chem Biol. 2020;27:420–35.
Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2020;66:89–100.
Bai YS, Meng LJ, Han L, Jia YY, Zhao YN, Gao H, et al. Lipid storage and lipophagy regulates ferroptosis. Biochem Bioph Res Co. 2019;508:997–1003.
Liu J, Yang M, Kang R, Klionsky DJ, Tang D. Autophagic degradation of the circadian clock regulator promotes ferroptosis. Autophagy. 2019;15:2033–5.
Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M, et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci USA. 2019;116:2996–3005.
Xue Q, Yan D, Chen X, Li X, Kang R, Klionsky DJ, et al. Copper-dependent autophagic degradation of GPX4 drives ferroptosis. Autophagy. 2023;19:1982–96.
Zhao L, Zhao J, Zhong K, Tong A, Jia D. Targeted protein degradation: mechanisms, strategies and application. Sig Transduct Target Ther. 2022;7:113.
Fan M, Gao J, Zhou L, Xue W, Wang Y, Chen J, et al. Highly expressed SERCA2 triggers tumor cell autophagy and is a druggable vulnerability in triple-negative breast cancer. Acta Pharm Sin B. 2022;12:4407–23.
Jiang T, Zhu J, Jiang S, Chen Z, Xu P, Gong R, et al. Targeting lncRNA DDIT4-AS1 Sensitizes Triple Negative Breast Cancer to Chemotherapy via Suppressing of Autophagy. Adv Sci. 2023;10:e2207257.
Guthrie RD Jr, Hines C Jr. Use of intravenous albumin in the critically ill patient. Am J Gastroenterol. 1991;86:255–63.
Hoogenboezem EN, Duvall CL. Harnessing albumin as a carrier for cancer therapies. Adv Drug Deliv Rev. 2018;130:73–89.
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82.
Baselga J, Bradbury I, Eidtmann H, Di Cosimo S, de Azambuja E, Aura C, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379:633–40.
Fuchs CS, Doi T, Jang RW, Muro K, Satoh T, Machado M, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018;4:e180013.
Kim DG, Kim M, Goo JI, Kong J, Harmalkar DS, Lu Q, et al. Chemical induction of the interaction between AIMP2-DX2 and Siah1 to enhance ubiquitination. Cell Chem Biol. 2024;31:1958–1968.e1958.
Zhong C, Zhu R, Jiang T, Tian S, Zhao X, Wan X, et al. Design and characterization of a novel eEF2K degrader with potent therapeutic efficacy against triple-negative breast cancer. Adv Sci (Weinh). 2024;11:e2305035.
Yang F, Xiao Y, Ding JH, Jin X, Ma D, Li DQ, et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023;35:84–100.e108.
Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10:1617.
Zhang HL, Hu BX, Ye ZP, Li ZL, Liu S, Zhong WQ, et al. TRPML1 triggers ferroptosis defense and is a potential therapeutic target in AKT-hyperactivated cancer. Sci Transl Med. 2024;16:eadk0330.
Xie Y, Kang R, Klionsky DJ, Tang D. GPX4 in cell death, autophagy, and disease. Autophagy. 2023;19:2621–38.
Funding
This study was funded by the National Natural Science Foundation of China (Project No. 82373064 and 81972480 for YC), Hunan Provincial Natural Science Foundation (Project No. 2022JJ80106 for YC), and the Project of Innovation-driven Plan in Central South University (Project No. 1053320221775 for XW).
Author information
Authors and Affiliations
Contributions
RG and XW contributed equally to this work. YC conceived and directed the project. RG and XW contributed to experimental design and program strategy. BX and JY contributed to design and chemical synthesis of compounds. RG and XW contributed to the in vitro biology experiments and data analysis. RG, XW, SJ, YG, YL, TJ, CZ, and LH contributed to the in vivo biology experiments and data analysis. ZY and ZC provided technological support in the experiments. RG and YC wrote the manuscript. JM and YC reviewed and edited the paper. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval
All the animal experiments were approved by the Ethics Committee of the Department of Laboratory Animals of Central South University (code/CSU-2022-0001-0029), and the animal protocol was in accordance with the institutional guidelines of the Animal Care and Use Committee of Central South University. The maximal tumor burden allowed by ethics did not exceed 2000 mm3. The ethical endpoint was determined once the maximal tumor burden was reached. All the experiments performed with human specimens were reviewed and approved by the Research Ethics Board at the Second Xiangya Hospital for Cancer Research (code/2023-0233). All patients whose tumor samples were used in the study gave informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gong, R., Wan, X., Jiang, S. et al. GPX4-AUTAC induces ferroptosis in breast cancer by promoting the selective autophagic degradation of GPX4 mediated by TRAF6-p62. Cell Death Differ 32, 2022–2037 (2025). https://doi.org/10.1038/s41418-025-01528-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-025-01528-1
This article is cited by
-
Targeted protein degradation: species, diseases and efficient utilization
Journal of Translational Medicine (2025)
-
Harnessing artificial intelligence to identify Bufalin as a molecular glue degrader of estrogen receptor alpha
Nature Communications (2025)
-
Inhibition of ferroptosis in inflammatory macrophages alleviates intestinal injury in neonatal necrotizing enterocolitis
Cell Death Discovery (2025)