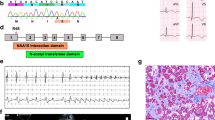
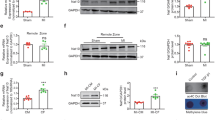
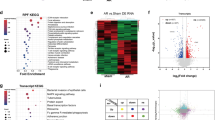
Abstract
Energy metabolism is crucial for heart development and function, and dysregulation of this process can lead to heart failure. However, the molecular mechanisms underlying these processes, particularly the role of RNA-binding proteins (RBPs)-mediated posttranscriptional regulation, remain largely unclear. We identified N-acetyltransferase 10 (NAT10) as a key regulator of heart function and cardiac diseases. NAT10 is crucial for heart development, and its dysregulation is associated with heart failure. Cardiac-specific deletion of Nat10 leads to dilated cardiomyopathy, heart failure, and postnatal death by downregulating genes related to fatty acid β-oxidation and heart contraction. Adult-onset knockout Nat10 also results in dilated cardiomyopathy and heart failure. NAT10-deficient hiPSC-CMs also showed impaired calcium transients during contraction. Restoration of NAT10(WT) and NAT10(G641E) (an N-acetyltransferase-inactive mutation), but not NAT10(K290A) (a loss-of-RNA-binding activity mutation), fully rescues the dilated cardiomyopathy, heart failure, and postnatal death phenotypes in Nat10-CKO mice by restoring expression of genes involved in fatty acid β-oxidation and heart contraction. The RNA-binding activity of NAT10 is essential for maintaining the expression of these genes. These findings demonstrate that NAT10 plays a critical role in heart development and function by maintaining the expression of genes related to fatty acid β-oxidation and heart contraction, highlighting its importance in maintaining heart health.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data are available in the main text or the supplementary materials. The RNA-seq data that support the findings of this study have been deposited in GEO under accession codes GSE274007, GSE274008, and GSE295332 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE274007; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE274008; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE295332). The RIP-seq data that support the findings of this study have been deposited in GEO under accession codes GSE274009 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE274009). The Ribo-seq data that support the findings of this study have been deposited in GEO under accession codes GSE295333 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE295333). All biological materials used are readily available from the authors or from standard commercial sources.
References
Sakamoto T, Kelly DP. Cardiac maturation. J Mol Cell Cardiol. 2024;187:38–50.
Zhou B, Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. J Clin Investig. 2018;128:3716–26.
Doenst T, Nguyen TD, Abel ED. Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res. 2013;113:709–24.
Alaynick WA, Kondo RP, Xie W, He W, Dufour CR, Downes M, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24.
Sakamoto T, Matsuura TR, Wan S, Ryba DM, Kim JU, Won KJ, et al. A critical role for estrogen-related receptor signaling in cardiac maturation. Circ Res. 2020;126:1685–702.
Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, et al. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat Med. 2004;10:1245–50.
Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–61.
Akerberg AA, Trembley M, Butty V, Schwertner A, Zhao L, Beerens M, et al. RBPMS2 is a myocardial-enriched splicing regulator required for cardiac function. Circ Res. 2022;131:980–1000.
Gan P, Wang Z, Morales MG, Zhang Y, Bassel-Duby R, Liu N, et al. RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev Cell. 2022;57:959–973.e957.
Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–86.e24.
Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, et al. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–66.
Liu X, Cai S, Zhang C, Liu Z, Luo J, Xing B, et al. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 2018;46:9601–16.
Wei W, Zhang S, Han H, Wang X, Zheng S, Wang Z, et al. NAT10-mediated ac4C tRNA modification promotes EGFR mRNA translation and gefitinib resistance in cancer. Cell Rep. 2023;42:112810.
Chen L, Wang WJ, Liu Q, Wu YK, Wu YW, Jiang Y, et al. NAT10-mediated N4-acetylcytidine modification is required for meiosis entry and progression in male germ cells. Nucleic Acids Res. 2022;50:10896–913.
Jiang X, Cheng Y, Zhu Y, Xu C, Li Q, Xing X, et al. Maternal NAT10 orchestrates oocyte meiotic cell-cycle progression and maturation in mice. Nat Commun. 2023;14:3729.
Liao L, He Y, Li SJ, Yu XM, Liu ZC, Liang YY, et al. Lysine 2-hydroxyisobutyrylation of NAT10 promotes cancer metastasis in an ac4C-dependent manner. Cell Res. 2023;33:355–71.
Balmus G, Larrieu D, Barros AC, Collins C, Abrudan M, Demir M, et al. Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat Commun. 2018;9:1700.
Shi J, Yang C, Zhang J, Zhao K, Li P, Kong C, et al. NAT10 is involved in cardiac remodeling through ac4C-mediated transcriptomic regulation. Circ Res. 2023;133:989–1002.
Xu T, Du T, Zhuang X, He X, Yan Y, Wu J, et al. Loss of NAT10 reduces the translation of Kmt5a mRNA through ac4C modification in cardiomyocytes and induces heart failure. J Am Heart Assoc. 2024;13:e035714.
Ma W, Tian Y, Shi L, Liang J, Ouyang Q, Li J, et al. N-acetyltransferase 10 represses Uqcr11 and Uqcrb independently of ac4C modification to promote heart regeneration. Nat Commun. 2024;15:2137.
Li X, Yuan B, Lu M, Wang Y, Ding N, Liu C, et al. The methyltransferase METTL3 negatively regulates nonalcoholic steatohepatitis (NASH) progression. Nat Commun. 2021;12:7213.
Li X, Ding K, Li X, Yuan B, Wang Y, Yao Z, et al. Deficiency of WTAP in hepatocytes induces lipoatrophy and non-alcoholic steatohepatitis (NASH). Nat Commun. 2022;13:4549.
Ding K, Zhang Z, Han Z, Shi L, Li X, Liu Y, et al. Liver ALKBH5 regulates glucose and lipid homeostasis independently through GCGR and mTORC1 signaling. Science. 2025;387:eadp4120.
Shi L, Li X, Zhang M, Qin C, Zhang Z, Chen Z. Downregulation of Wtap causes dilated cardiomyopathy and heart failure. J Mol Cell Cardiol. 2024;188:38–51.
Vandergriff AC, Hensley MT, Cheng K. Isolation and cryopreservation of neonatal rat cardiomyocytes. J Vis Exp. 2015;98:52726.
Liu R, Wubulikasimu Z, Cai R, Meng F, Cui Q, Zhou Y, et al. NAT10-mediated N4-acetylcytidine mRNA modification regulates self-renewal in human embryonic stem cells. Nucleic Acids Res. 2023;51:8514–31.
Zhang JZ, Belbachir N, Zhang T, Liu Y, Shrestha R, Wu JC. Effects of cryopreservation on human induced pluripotent stem cell-derived cardiomyocytes for assessing drug safety response profiles. Stem Cell Rep. 2021;16:168–81.
Yang H, Yang Y, Lu Z, Zhang JZ. Simultaneous optical imaging of action potentials and calcium transients in human induced pluripotent stem cell-derived cardiomyocytes. Curr Protoc. 2024;4:e1101.
Li X, Jia L, Chen X, Dong Y, Ren X, Dong Y, et al. Islet α-cell inflammation induced by NF-κB inducing kinase (NIK) leads to hypoglycemia, pancreatitis, growth retardation, and postnatal death in mice. Theranostics. 2018;8:5960–71.
Jia L, Jiang Y, Li X, Chen Z. Purbeta promotes hepatic glucose production by increasing Adcy6 transcription. Mol Metab. 2020;31:85–97.
Wang L, Wang Y, Ding K, Li Z, Zhang Z, Li X, et al. YTHDC1 promotes postnatal brown adipose tissue development and thermogenesis by stabilizing PPARγ. EMBO J. 2025;44:3360–80.
Li X, Li X, Liu C, Li Z, Ding K, Wang Y, et al. YTHDC1 is essential for postnatal liver development and homeostasis. Adv Sci. 2025:e05725.
Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33:722–9.
Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–50.
Lauria F, Tebaldi T, Bernabò P, Groen EJN, Gillingwater TH, Viero G. riboWaltz: Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput. Biol. 2018;14:e1006169.
Chothani S, Adami E, Ouyang JF, Viswanathan S, Hubner N, Cook SA, et al. deltaTE: detection of translationally regulated genes by integrative analysis of ribo-seq and RNA-seq data. Curr Protoc Mol Biol. 2019;129:e108.
Nomura S, Satoh M, Fujita T, Higo T, Sumida T, Ko T, et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun. 2018;9:4435.
Sweet ME, Cocciolo A, Slavov D, Jones KL, Sweet JR, Graw SL, et al. Transcriptome analysis of human heart failure reveals dysregulated cell adhesion in dilated cardiomyopathy and activated immune pathways in ischemic heart failure. BMC Genom. 2018;19:812.
Werfel S, Jungmann A, Lehmann L, Ksienzyk J, Bekeredjian R, Kaya Z, et al. Rapid and highly efficient inducible cardiac gene knockout in adult mice using AAV-mediated expression of Cre recombinase. Cardiovasc Res. 2014;104:15–23.
Wei R, Cui X, Min J, Lin Z, Zhou Y, Guo M, et al. NAT10 promotes cell proliferation by acetylating CEP170 mRNA to enhance translation efficiency in multiple myeloma. Acta Pharm Sin B. 2022;12:3313–25.
Reichart D, Lindberg EL, Maatz H, Miranda AMA, Viveiros A, Shvetsov N, et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science. 2022;377:eabo1984.
Chaffin M, Papangeli I, Simonson B, Akkad AD, Hill MC, Arduini A, et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy. Nature. 2022;608:174–80.
Qu Z, Pang X, Mei Z, Li Y, Zhang Y, Huang C, et al. The positive feedback loop of the NAT10/Mybbp1a/p53 axis promotes cardiomyocyte ferroptosis to exacerbate cardiac I/R injury. Redox Biol. 2024;72:103145.
Tsai K, Jaguva Vasudevan AA, Martinez Campos C, Emery A, Swanstrom R, Cullen BR. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe. 2020;28:306–312.e6.
Yu C, Chen Y, Luo H, Lin W, Lin X, Jiang Q, et al. NAT10 promotes vascular remodelling via mRNA ac4C acetylation. Eur Heart J. 2024;46:288–304.
Acknowledgements
We thank Novogene and DIATRE Biotechnology for assistance in RNA-seq, RIP-seq and Ribo-seq experiments, respectively.
Funding
This study was supported by the National Natural Science Foundation of China Grants (82525015, 32371198 and 82370861), the Heilongjiang Chunyan Team (CYCX24020), Frontiers Science Center for Matter Behave in Space Environment and the Fundamental Research Funds for the Central Universities (HIT.OCEF.2025001), Doctor of excellence program (DEP) at the First Hospital of Jilin University (JDYY-DEP-2024040).
Author information
Authors and Affiliations
Contributions
LS performed most of the experiments and analyzed the data. MZ performed the experiments. HY, SH and JZ performed the iPSC-CM experiments and analyzed the data. XL performed RIP-seq experiments. YC helped to analyze the RNA-seq data. MG provided Nat10flox/flox mice and reviewed this study. ZZ and ZL guided this study. ZC conceived and designed the project, researched data, and wrote the manuscript. All authors give their consent for the publication of the above manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Institutional Animal Care and Use Committee or Animal Experimental Ethics Committee of Harbin Institute of Technology (HIT/IACUC-2022066). All authors complied with all relevant ethical regulations for animal testing and research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, L., Zhang, M., Yang, H. et al. NAT10 regulates heart development and function by maintaining the expression of genes related to fatty acid β-oxidation and heart contraction. Cell Death Differ (2025). https://doi.org/10.1038/s41418-025-01577-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-025-01577-6