Abstract
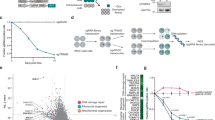
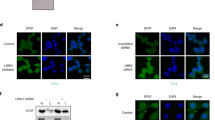
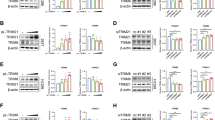
SPOP, the most frequently mutated gene in prostate cancer, has been implicated in the aberrant activation of stress granules, presenting significant challenges in disease management. However, the mechanistic link between SPOP mutations and cellular energy stress remains inadequately explored. In this study, we demonstrate that ULK1 expression is positively correlated with both loss-of-function mutations in SPOP and the upregulation of the E3 ubiquitin ligase TRIM24 in human prostate cancer specimens. Mechanistically, SPOP mutations induce the upregulation of TRIM24, which subsequently binds to ULK1 and catalyzes its non-degradative K27-linked polyubiquitylation. This post-translational modification enhances the stability of ULK1, facilitating cellular adaptation to energy stress and consequently promoting prostate cancer progression. Notably, pharmacological inhibition of TRIM24 using TRIM24-PROTAC (proteolysis-targeting chimera) effectively suppressed tumor growth in mice bearing SPOP-mutant prostate cancer cells. Collectively, these findings elucidate a pivotal role of SPOP mutations in modulating energy stress responses via TRIM24-mediated ULK1 ubiquitylation and underscore the therapeutic potential of targeting TRIM24 in SPOP-mutant prostate cancers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Huang J, Chan EO, Liu X, Lok V, Ngai CH, Zhang L, et al. Global trends of prostate cancer by age, and their associations with gross domestic product (GDP), human development index (HDI), smoking, and alcohol drinking. Clin Genitourin Cancer. 2023;21:e261–70.e50.
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48.
Zhang Y, Chen XN, Zhang H, Wen JK, Gao HT, Shi B, et al. CDK13 promotes lipid deposition and prostate cancer progression by stimulating NSUN5-mediated m5C modification of ACC1 mRNA. Cell Death Differ. 2023;30:2462–76.
Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, Tannock I, et al. Prostate cancer. Lancet. 2016;387:70–82.
Li J, Zhang T, Ren T, Liao X, Hao Y, Lim JS, et al. Oxygen-sensitive methylation of ULK1 is required for hypoxia-induced autophagy. Nat Commun. 2022;13:1172.
Xiao M, Zhao J, Wang Q, Liu J, Ma L. Recent advances of degradation technologies based on PROTAC mechanism. Biomolecules. 2022;12:1257.
Rebello RJ, Oing C, Knudsen KE, Loeb S, Johnson DC, Reiter RE, et al. Prostate cancer. Nat Rev Dis Prim. 2021;7:9.
Song Y, Xu Y, Pan C, Yan L, Wang ZW, Zhu X. The emerging role of SPOP protein in tumorigenesis and cancer therapy. Mol Cancer. 2020;19:2.
Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50.
Marzahn MR, Marada S, Lee J, Nourse A, Kenrick S, Zhao H, et al. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016;35:1254–75.
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–25.
Jiang Q, Zheng N, Bu L, Zhang X, Zhang X, Wu Y, et al. SPOP-mediated ubiquitination and degradation of PDK1 suppresses AKT kinase activity and oncogenic functions. Mol Cancer. 2021;20:100.
Ding G, Sun J, Jiang L, Gao P, Zhou Q, Wang J, et al. Key pathways in prostate cancer with SPOP mutation identified by bioinformatic analysis. Open Med. 2020;15:1039–47.
An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–69.
Wang Z, Song Y, Ye M, Dai X, Zhu X, Wei W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol. 2020;17:339–50.
Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med. 2017;23:1055–62.
Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–5.
Zhu H, Ren S, Bitler BG, Aird KM, Tu Z, Skordalakes E, et al. SPOP E3 ubiquitin ligase adaptor promotes cellular senescence by degrading the SENP7 deSUMOylase. Cell Rep. 2015;13:1183–93.
Shi Q, Zhu Y, Ma J, Chang K, Ding D, Bai Y, et al. Prostate cancer-associated SPOP mutations enhance cancer cell survival and docetaxel resistance by upregulating Caprin1-dependent stress granule assembly. Mol Cancer. 2019;18:170.
Zhang J, Chen M, Zhu Y, Dai X, Dang F, Ren J, et al. SPOP promotes nanog destruction to suppress stem cell traits and prostate cancer progression. Dev Cell. 2019;48:329–44.e5.
Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–30.
Wu Q, Tian AL, Durand S, Aprahamian F, Nirmalathasan N, Xie W, et al. Isobacachalcone induces autophagy and improves the outcome of immunogenic chemotherapy. Cell Death Dis. 2020;11:1015.
Wu Q, Tian AL, Kroemer G, Kepp O. Autophagy induction by IGF1R inhibition with picropodophyllin and linsitinib. Autophagy. 2021;17:2046–7.
Blessing AM, Rajapakshe K, Reddy Bollu L, Shi Y, White MA, Pham AH, et al. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy. 2017;13:506–21.
Ashrafizadeh M, Paskeh MDA, Mirzaei S, Gholami MH, Zarrabi A, Hashemi F, et al. Targeting autophagy in prostate cancer: preclinical and clinical evidence for therapeutic response. J Exp Clin Cancer Res. 2022;41:105.
Farrow JM, Yang JC, Evans CP. Autophagy as a modulator and target in prostate cancer. Nat Rev Urol. 2014;11:508–16.
Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–15.
Zachari M, Ganley IG. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61:585–96.
Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–62.
Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol. 2013;20:144–9.
Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50.
Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41.
Alers S, Löffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11.
Yang Y, Zhu Y, Zhou S, Tang P, Xu R, Zhang Y, et al. TRIM27 cooperates with STK38L to inhibit ULK1-mediated autophagy and promote tumorigenesis. EMBO J. 2022;41:e109777.
Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61:84–97.
Gao K, Shi Q, Liu Y, Wang C. Enhanced autophagy and NFE2L2/NRF2 pathway activation in SPOP mutation-driven prostate cancer. Autophagy. 2022;18:2013–5.
Shi Q, Jin X, Zhang P, Li Q, Lv Z, Ding Y, et al. SPOP mutations promote p62/SQSTM1-dependent autophagy and Nrf2 activation in prostate cancer. Cell Death Differ. 2022;29:1228–39.
Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846–58.
Gechijian LN, Buckley DL, Lawlor MA, Reyes JM, Paulk J, Ott CJ, et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat Chem Biol. 2018;14:405–12.
Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci USA. 2013;110:6997–7002.
Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74:5631–43.
Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346:85–9.
Feng Y, Klionsky DJ. Downregulation of autophagy through CUL3-KLHL20-mediated turnover of the ULK1 and PIK3C3/VPS34 complexes. Autophagy. 2016;12:1071–2.
Wei X, Fried J, Li Y, Hu L, Gao M, Zhang S, et al. Functional roles of speckle-type poz (SPOP) protein in genomic stability. J Cancer. 2018;9:3257–62.
Fan Y, Hou T, Dan W, Zhu Y, Liu B, Wei Y, et al. ERK1/2 inhibits Cullin 3/SPOP-mediated PrLZ ubiquitination and degradation to modulate prostate cancer progression. Cell Death Differ. 2022;29:1611–24.
Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA. 2001;98:8554–9.
Burke MR, Smith AR, Zheng G. Overcoming cancer drug resistance utilizing PROTAC technology. Front Cell Dev Biol. 2022;10:872729.
Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. 2022;21:181–200.
Chen X, Shen H, Shao Y, Ma Q, Niu Y, Shang Z. A narrative review of proteolytic targeting chimeras (PROTACs): future perspective for prostate cancer therapy. Transl Androl Urol. 2021;10:954–62.
Jia X, Han X. Targeting androgen receptor degradation with PROTACs from bench to bedside. Biomed Pharmacother. 2023;158:114112.
Yan W, Shi X, Wang H, Liao A, Yang W. Aberrant SPOP-CHAF1A ubiquitination axis triggers tumor autophagy that endows a therapeutical vulnerability in diffuse large B cell lymphoma. J Transl Med. 2022;20:296.
Jamali L, Moradi A, Ganji M, Ayati M, Kazeminezhad B, Fazeli Attar Z, et al. Potential prognostic role for SPOP, DAXX, RARRES1, and LAMP2 as an autophagy related genes in prostate cancer. Urol J. 2020;17:156–63.
Guo D, Yu Q, Tong Y, Qian X, Meng Y, Ye F, et al. OXCT1 succinylation and activation by SUCLA2 promotes ketolysis and liver tumor growth. Mol Cell. 2025;85:843–56.e6.
Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med. 2017;23:1063–71.
Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, et al. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol Cell. 2015;59:917–30.
Acknowledgements
The authors would like to express sincere gratitude to Cosmos Wisdom Company for their generous support and invaluable assistance throughout this research. The company’s technical support significantly contributed to the completion of this study. We deeply appreciate them for their support.
Funding
This work was supported by the National Natural Science Foundation of China (82372806 to LM, 82203450 to YZ, 82125026 to HR, 82472667 to ZY, 82188102 and 82030074 to ZL), the Ministry of Science and Technology of the People’s Republic of China (2020YFA0803300, ZL), the Natural Science Foundation of Shandong Province, China (ZR2021MC039 to LM), the Taishan Scholar Foundation of Shandong Province (tsqn202312378 to QW), Taishan Scholars Young Experts Project (tsqn202312380 to ZY) and Qingdao Key Clinical Specialty Elite Discipline. This work was supported by the Shanghai Sailing Program to YZ (20YF1448100 to YZ) and grants from the Science and Technology Commission of Shanghai Municipality (22Y11905200 to YZ).
Author information
Authors and Affiliations
Contributions
LM, YZ, DW, JLiu, QW, ZL, and HR. conceived and designed the study; LM, JLin, DW, QW, ZY, QL, SC, XL, WF, JZ, SY, ZW, XQ, YW, YQ, YC, and YX performed the experiments; JLiu, DW, YZ, and JLin performed the statistical analysis and wrote the original draft. LM, YZ, and HR contributed to the final draft. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Institute Review Board of Shanghai Changhai Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, S., Lin, J., Yang, Z. et al. TRIM24-mediated K27-linked ubiquitination of ULK1 alleviates energy stress-induced autophagy and promote prostate cancer growth in the context of SPOP mutation. Cell Death Differ (2025). https://doi.org/10.1038/s41418-025-01582-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41418-025-01582-9