Abstract
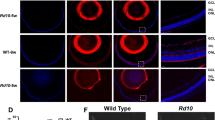
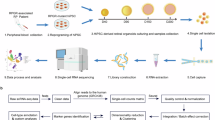
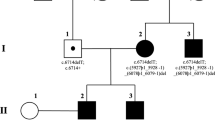
Retinitis pigmentosa (RP), affecting more than 20 million people worldwide, refers to a group of inherited retinal dystrophies characterized by progressive photoreceptor degeneration and vision loss. However, the underlying genetic causes of substantial RP cases remain unidentified. In this report, we identified a novel homozygous splicing variant, c.219-1delG, which introduced skipping of exon 4 of the ZNF124 gene in a large RP pedigree by whole-exome sequencing analysis. To elucidate the pathogenesis of the mutation, we generated a retina-specific knockout mouse model of ZNF124 murine homologous gene Gm20541, which manifested RP-like phenotypes characterized by reduced electroretinogram response and progressive retinal degeneration. Integrated analysis using CUT&Tag, ChIP-exo, and RNA-seq data further revealed that ZNF124 regulated MSX2 expression through binding its promoter region. Moreover, deletion of Msx2in the retina led to thinning of retina owing to progressive degeneration of rod cells. Integrated analysis of RNA-seq data from both Gm20541 and Msx2 mutant retinas indicated that ZNF124 is essential for maintaining normal retinal function by regulating Msx2 transcription, which in turn controls the expression of murine homologues of retinal dystrophy genes Rs1, Pde6g, and Pdc. Taken together, our study identified a novel mechanism of transcriptional regulation for retinal homeostasis via ZNF124-MSX2 axis and ZNF124 as a novel candidate gene for RP.
Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–809.
Chang S, Vaccarella L, Olatunji S, Cebulla C, Christoforidis J. Diagnostic challenges in retinitis pigmentosa: genotypic multiplicity and phenotypic variability. Curr Genom. 2011;12:267–75.
Narayan DS, Wood JP, Chidlow G, Casson RJ. A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmol. 2016;94:748–54.
Daiger S, Sullivan L, Bowne S. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–41.
Sandberg MA, Rosner B, Weigel-DiFranco C, Dryja TP, Berson EL. Disease course of patients with X-linked retinitis pigmentosa due to RPGR gene mutations. Investig Ophthalmol Vis Sci. 2007;48:1298–304.
Neveling K, Collin RW, Gilissen C, van Huet RA, Visser L, Kwint MP, et al. Next-generation genetic testing for retinitis pigmentosa. Hum Mutat. 2012;33:963–72.
Thomas JH, Emerson RO. Evolution of C2H2-zinc finger genes revisited. BMC Evolut Biol. 2009;9:1–8.
Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–77.
Yang P, Wang Y, Macfarlan TS. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. 2017;33:871–81.
Kuramoto K, Uesaka T, Kimura A, Kobayashi M, Watanabe H, Katoh O. ZK7, a novel zinc finger gene, is induced by vascular endothelial growth factor and inhibits apoptotic death in hematopoietic cells. Cancer Res. 2000;60:425–30.
Miyake N, Katoh O, Hirata S, Kimura S, Watanabe H, Yajin K. Expression of the Krüppel-type zinc finger gene, ZK7, in head and neck squamous cell carcinoma and normal mucosa. Cancer Lett. 2002;185:111–8.
Luo Z, Dong X, Yu J, Xia Y, Berry KP, Rao R, et al. Genomic and transcriptomic analyses reveals ZNF124 as a critical regulator in highly aggressive medulloblastomas. Front Cell Dev Biol. 2021;9:634056.
Poot M, Kroes HY, V D Wijst SE, Eleveld MJ, Rooms L, Nievelstein RAJ, et al. Dandy-Walker complex in a boy with a 5 Mb deletion of region 1q44 due to a paternal t (1; 20)(q44; q13. 33). Am J Med Genet Part A. 2007;143:1038–44.
Han J, Ishii M, Bringas P Jr, Maas RL, Maxson RE Jr, Chai Y. Concerted action of Msx1 and Msx2 in regulating cranial neural crest cell differentiation during frontal bone development. Mech Dev. 2007;124:729–45.
Antonopoulou I, Mavrogiannis LA, Wilkie AO, Morriss-Kay GM. Alx4 and Msx2 play phenotypically similar and additive roles in skull vault differentiation. J Anat. 2004;204:487–99.
Zhang Q, Huang Z, Zuo H, Lin Y, Xiao Y, Yan Y, et al. Chromatin accessibility predetermines odontoblast terminal differentiation. Front Cell Dev Biol. 2021;9:769193.
Wu L-Y, Li M, Hinton DR, Guo L, Jiang S, Wang JT, et al. Microphthalmia resulting from MSX2-induced apoptosis in the optic vesicle. Investig Ophthalmol Vis Sci. 2003;44:2404–12.
Foerst-Potts L, Sadler T. Disruption of Msx-1 and Msx-2 reveals roles for these genes in craniofacial, eye, and axial development. Dev Dyn Off Publ Am Assoc Anat. 1997;209:70–84.
Jiang S-Y, Wang J-T. Msx2 alters the timing of retinal ganglion cells fate commitment and differentiation. Biochem Biophys Res Commun. 2010;395:524–9.
Zhao J, Kawai K, Wang H, Wu D, Wang M, Yue Z, et al. Loss of Msx2 function down-regulates the FoxE3 expression and results in anterior segment dysgenesis resembling Peters anomaly. Am J Pathol. 2012;180:2230–9.
Yu Z, Yu W, Liu J, Wu D, Wang C, Zhang J, et al. Lens-specific deletion of the Msx2 gene increased apoptosis by enhancing the caspase-3/caspase-8 signaling pathway. J Int Med Res. 2018;46:2843–55.
Zhou Y, Li S, Huang L, Yang Y, Zhang L, Yang M, et al. A splicing mutation in aryl hydrocarbon receptor associated with retinitis pigmentosa. Hum Mol Genet. 2018;27:2563–72.
Zhang L, Sun Z, Zhao P, Huang L, Xu M, Yang Y, et al. Whole-exome sequencing revealed HKDC1 as a candidate gene associated with autosomal-recessive retinitis pigmentosa. Hum Mol Genet. 2018;27:4157–68.
Corrionero A, Raker VA, Izquierdo JM, Valcárcel J. Strict 3′ splice site sequence requirements for U2 snRNP recruitment after U2AF binding underlie a genetic defect leading to autoimmune disease. RNA. 2011;17:401–11.
Clancy S. RNA splicing: introns, exons and spliceosome. Nat Educ. 2008;1:31.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419.
Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. genesis. 2000;26:130–2.
Eldred KC, Avelis C, Johnston RJ Jr, Roberts E. Modeling binary and graded cone cell fate patterning in the mouse retina. PLoS Comput Biol. 2020;16:e1007691.
Vidal L, Díaz F, Villena A, Moreno M, Campos JG, Pérez de Vargas I. Reaction of Müller cells in an experimental rat model of increased intraocular pressure following timolol, latanoprost and brimonidine. Brain Res Bull. 2010;82:18–24.
Yang Y, Shuai P, Li X, Sun K, Jiang X, Liu W, et al. Mettl14-mediated m6A modification is essential for visual function and retinal photoreceptor survival. BMC Biol. 2022;20:140.
Imbeault M, Helleboid P-Y, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543:550–4.
Plaisancié J, Collet C, Pelletier V, Perdomo Y, Studer F, Fradin M, et al. MSX2 gene duplication in a patient with eye development defects. Ophthalmic Genet. 2015;36:353–8.
Markitantova Y, Simirskii V. Inherited eye diseases with retinal manifestations through the eyes of homeobox genes. Int J Mol Sci. 2020;21:1602.
Bujakowska K, Audo I, Mohand-Saïd S, Lancelot ME, Antonio A, Germain A, et al. CRB1 mutations in inherited retinal dystrophies. Hum Mutat. 2012;33:306–15.
den Hollander AI, Davis J, van der Velde-Visser SD, Zonneveld MN, Pierrottet CO, Koenekoop RK, et al. CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat. 2004;24:355–69.
den Hollander AI, Heckenlively JR, van den Born LI, de Kok YJ, van der Velde-Visser SD, Kellner U, et al. Leber congenital amaurosis and retinitis pigmentosa with Coats-like exudative vasculopathy are associated with mutations in the crumbs homologue 1 (CRB1) gene. Am J Hum Genet. 2001;69:198–203.
Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, et al. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–89.
Aleman TS, Cideciyan AV, Aguirre GK, Huang WC, Mullins CL, Roman AJ, et al. Human CRB1-associated retinal degeneration: comparison with the rd8 Crb1-mutant mouse model. Invest Ophthalmol Vis Sci. 2011;52:6898–910.
Luhmann UF, Carvalho LS, Holthaus SM, Cowing JA, Greenaway S, Chu CJ, et al. The severity of retinal pathology in homozygous Crb1rd8/rd8 mice is dependent on additional genetic factors. Hum Mol Genet. 2015;24:128–41.
Jones BW, Marc RE. Retinal remodeling during retinal degeneration. Exp Eye Res. 2005;81:123–37.
Strettoi E, Pignatelli V, Rossi C, Porciatti V, Falsini B. Remodeling of second-order neurons in the retina of rd/rd mutant mice. Vis Res. 2003;43:867–77.
Ekker M, Akimenko MA, Allende ML, Smith R, Drouin G, Langille RM, et al. Relationships among msx gene structure and function in zebrafish and other vertebrates. Mol Biol Evol. 1997;14:1008–22.
Tolun G, Vijayasarathy C, Huang R, Zeng Y, Li Y, Steven AC, et al. Paired octamer rings of retinoschisin suggest a junctional model for cell–cell adhesion in the retina. Proc Natl Acad Sci. 2016;113:5287–92.
Heymann JB, Vijayasarathy C, Huang RK, Dearborn AD, Sieving PA, Steven AC. Cryo-EM of retinoschisin branched networks suggests an intercellular adhesive scaffold in the retina. J Cell Biol. 2019;218:1027–38.
Molday RS, Kellner U, Weber BH. X-linked juvenile retinoschisis: clinical diagnosis, genetic analysis, and molecular mechanisms. Prog Retin Eye Res. 2012;31:195–212.
Vijayasarathy C, Pasha SPBS, Sieving PA. Of men and mice: Human X-linked retinoschisis and fidelity in mouse modeling. Prog Retin Eye Res. 2022;87:100999.
Baehr W, Devlin MJ, Applebury ML. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J Biol Chem. 1979;254:11669–77.
Hurley J, Stryer L. Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J Biol Chem. 1982;257:11094–9.
Farber DB, Tsang SH. Stationary night blindness or progressive retinal degeneration in mice carrying different alleles of PDE gamma. Front Biosci. 2003;8:s666–s675.
Nishiguchi KM, Berson EL, Dryja TP. Mutation screening of the phosducin gene PDC in patients with retinitis pigmentosa and allied diseases. Mol Vis. 2004;10:62–4.
Lee RH, Whelan JP, Lolley RN, McGinnis JF. The photoreceptor-specific 33 kDa phosphoprotein of mammalian retina: generation of monospecific antibodies and localization by immunocytochemistry. Exp Eye Res. 1988;46:829–40.
Sokolov M, Strissel KJ, Leskov IB, Michaud NA, Govardovskii VI, Arshavsky VY. Phosducin facilitates light-driven transducin translocation in rod photoreceptors: evidence from the phosducin knockout mouse. J Biol Chem. 2004;279:19149–56.
Krispel CM, Sokolov M, Chen YM, Song H, Herrmann R, Arshavsky VY, et al. Phosducin regulates the expression of transducin βγ subunits in rod photoreceptors and does not contribute to phototransduction adaptation. J Gen Physiol. 2007;130:303–12.
Yang Y, Li X, Wang J, Tan J, Fitzmaurice B, Nishina PM, et al. A missense mutation in Pitx2 leads to early-onset glaucoma via NRF2-YAP1 axis. Cell Death Dis. 2021;12:1017.
Henikoff S, Henikoff JG, Kaya-Okur HS, Ahmad K. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930.
Acknowledgements
This study was supported by the National Key R&D Program of China (grant no. 2024YFC2510900/2024YFC2510902), the National Natural Science Foundation of China (82371083, 82471100, 82121003, 82271084), the Department of Science and Technology of Sichuan Province (2023ZYD0172, 23ZDYF2057), research grant from Jinfeng Laboratory (JFLKYXM202403AZ-101). The funders had no role in the study design, data collection and analysis, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
XZ, ZY, LZ, LH, and PS conceived the project. XZ and ZY designed experiments. YY, XJ, SL, KS, RZ, CC, YS, and WL acquired data. YY, LH, and XZ analyzed data. YY, PS and XZ interpreted data and drafted the manuscript. XZ, ZY, LZ, and SL revised the manuscript. All authors approved submission for publication.
Corresponding authors
Ethics declarations
Competing interests
All authors declare that there are no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Wei-Na Jin
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Jiang, X., Li, S. et al. Unveiling ZNF124 as a novel determinant in neurodegeneration: orchestration of photoreceptor homeostasis through MSX2 transcriptional regulation. Cell Death Dis (2026). https://doi.org/10.1038/s41419-026-08487-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-026-08487-6