Abstract
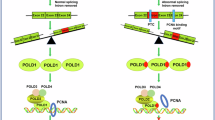
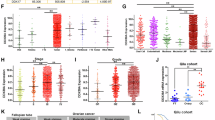
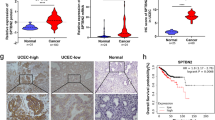
Recent studies have reported the overexpression of Sm proteins in several cancers, suggesting their potential as therapeutic targets; however, the specific Sm family members involved in endometrial cancer and their mechanisms remain unclear. Here, we show that the Sm protein SNRPD2 is markedly upregulated in both fresh-frozen and formalin-fixed paraffin-embedded (FFPE) endometrial cancer specimens and that its overexpression correlates with poorer clinical outcomes. In vitro and in vivo functional assays demonstrate that silencing SNRPD2 suppresses endometrial cancer cell proliferation and metastasis. Specifically, antisense oligonucleotides (ASOs) targeting SNRPD2 markedly reduced tumor growth in a patient-derived xenograft (PDX) model. Mechanistic analyses reveal that SNRPD2 knockdown induces the retention of intron 5 in DDX39B, resulting in the production of a noncoding transcript that is degraded by the nonsense-mediated decay (NMD) pathway and thereby decreases DDX39B expression. Reduced DDX39B levels permit the activation of a cryptic exon (Exon 2_3) in the CTSC mRNA, which introduces premature termination codons (PTCs) and triggers additional NMD-mediated degradation, leading to decreased CTSC expression. Thus, SNRPD2 maintains high DDX39B expression by preventing intron retention, and in turn, elevated DDX39B expression suppresses cryptic exon usage in CTSC to preserve CTSC expression, ultimately supporting malignant phenotypes of endometrial cancer. These results define a novel SNRPD2–DDX39B–CTSC regulatory axis and identify SNRPD2 as a promising therapeutic target for endometrial cancer.
Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Makker V, MacKay H, Ray-Coquard I, Levine DA, Westin SN, Aoki D, et al. Endometrial cancer. Nat Rev Dis Prim. 2021;7:88.
Salmon A, Lebeau A, Streel S, Dheur A, Schoenen S, Goffin F, et al. Locally advanced and metastatic endometrial cancer: current and emerging therapies. Cancer Treat Rev. 2024;129:102790.
Njoku K, Pierce A, Chiasserini D, Geary B, Campbell AE, Kelsall J, et al. Detection of endometrial cancer in cervico-vaginal fluid and blood plasma: leveraging proteomics and machine learning for biomarker discovery. EBioMedicine. 2024;102:105064.
Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025;75:10–45.
Sheikh MA, Althouse AD, Freese KE, Soisson S, Edwards RP, Welburn S, et al. USA endometrial cancer projections to 2030: should we be concerned?. Future Oncol. 2014;10:2561–8.
Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–21.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Corr BR, Erickson BK, Barber EL, Fisher CM, Slomovitz B. Advances in the management of endometrial cancer. BMJ. 2025;388:e080978.
Zhang Q, Ai Y, Abdel-Wahab O. Molecular impact of mutations in RNA splicing factors in cancer. Mol Cell. 2024;84:3667–80.
Bradley RK, Anczukow O. RNA splicing dysregulation and the hallmarks of cancer. Nat Rev Cancer. 2023;23:135–55.
Wu Q, Liao R, Miao C, Hasnat M, Li L, Sun L, et al. Oncofetal SNRPE promotes HCC tumorigenesis by regulating the FGFR4 expression through alternative splicing. Br J Cancer. 2024;131:77–89.
Yang H, Beutler B, Zhang D. Emerging roles of spliceosome in cancer and immunity. Protein Cell. 2022;13:559–79.
Wei L, Li Y, Chen J, Wang Y, Wu J, Yang H, et al. Alternative splicing in ovarian cancer. Cell Commun Signal. 2024;22:507.
Li Y, Chen Z, Peng J, Yuan C, Yan S, Yang N, et al. The splicing factor SNRPB promotes ovarian cancer progression through regulating aberrant exon skipping of POLA1 and BRCA2. Oncogene. 2023;42:2386–401.
Lu XX, Yang WX, Pei YC, Luo H, Li XG, Wang YJ, et al. An in vivo CRISPR screen identifies that SNRPC promotes triple-negative breast cancer progression. Cancer Res. 2023;83:2000–15.
Dai X, Yu L, Chen X, Zhang J. SNRPD1 confers diagnostic and therapeutic values on breast cancers through cell cycle regulation. Cancer Cell Int. 2021;21:229.
Salib A, Jayatilleke N, Seneviratne JA, Mayoh C, De Preter K, Speleman F, et al. MYCN and SNRPD3 cooperate to maintain a balance of alternative splicing events that drives neuroblastoma progression. Oncogene. 2024;43:363–77.
Sun L, Liu Y, Guo X, Cui T, Wu C, Tao J, et al. Acetylation-dependent regulation of core spliceosome modulates hepatocellular carcinoma cassette exons and sensitivity to PARP inhibitors. Nat Commun. 2024;15:5209.
Chang C, Li L, Su L, Yang F, Zha Q, Sun M, et al. Intron retention of DDX39A driven by SNRPD2 is a crucial splicing axis for oncogenic MYC/spliceosome program in hepatocellular carcinoma. Adv Sci. 2024;11:e2403387.
Koedoot E, van Steijn E, Vermeer M, Gonzalez-Prieto R, Vertegaal ACO, Martens JWM, et al. Splicing factors control triple-negative breast cancer cell mitosis through SUN2 interaction and sororin intron retention. J Exp Clin Cancer Res. 2021;40:82.
Hu Z, Li M, Chen Y, Chen L, Han Y, Chen C, et al. Disruption of PABPN1 phase separation by SNRPD2 drives colorectal cancer cell proliferation and migration through promoting alternative polyadenylation of CTNNBIP1. Sci China Life Sci. 2024;67:1212–25.
Li J, Li P, Brachtlova T, van der Meulen-Muileman IH, Dekker H, Kumar VS, et al. Evaluation of spliceosome protein SmD2 as a potential target for cancer therapy. Int J Mol Sci. 2024;25:13131.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60.
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8.
Whiteaker JR, Halusa GN, Hoofnagle AN, Sharma V, MacLean B, Yan P, et al. CPTAC Assay Portal: a repository of targeted proteomic assays. Nat Methods. 2014;11:703–4.
Nagy A, Munkacsy G, Gyorffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11:6047.
Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27.
Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50:W216–21.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202.
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601.
Li Y, Chen Z, Xiao H, Liu Y, Zhao C, Yang N, et al. Targeting the splicing factor SNRPB inhibits endometrial cancer progression by retaining the POLD1 intron. Exp Mol Med. 2025;57:420–35.
Li Y, Chen Z, Gao Y, Liu Y, Gao Q, Pu Y, et al. SNRPB-mediated regulation of DDX39A splicing promotes ovarian cancer progression by regulating alpha6 integrin subunit expression. Oncogene. 2025;44:2170–85.
Banerjee S, Nagasawa CK, Widen SG, Garcia-Blanco MA. Parsing the roles of DExD-box proteins DDX39A and DDX39B in alternative RNA splicing. Nucleic Acids Res. 2024;52:8534–51.
Zhang H, He C, Guo X, Fang Y, Lai Q, Wang X, et al. DDX39B contributes to the proliferation of colorectal cancer through direct binding to CDK6/CCND1. Cell Death Discov. 2022;8:30.
Hirano M, Galarza-Munoz G, Nagasawa C, Schott G, Wang L, Antonia AL, et al. The RNA helicase DDX39B activates FOXP3 RNA splicing to control T regulatory cell fate. eLife. 2023;12:e76927.
Li Q, Yuan H, Zhao G, Ou D, Zhang J, Li L, et al. DDX39B protects against sorafenib-induced ferroptosis by facilitating the splicing and cytoplasmic export of GPX4 pre-mRNA in hepatocellular carcinoma. Biochem Pharm. 2024;225:116251.
He C, Li A, Lai Q, Ding J, Yan Q, Liu S, et al. The DDX39B/FUT3/TGFbetaR-I axis promotes tumor metastasis and EMT in colorectal cancer. Cell Death Dis. 2021;12:74.
Khaket TP, Singh MP, Khan I, Bhardwaj M, Kang SC. Targeting of cathepsin C induces autophagic dysregulation that directs ER stress mediated cellular cytotoxicity in colorectal cancer cells. Cell Signal. 2018;46:92–102.
Wang X, Jia Y, Wang D. Cathepsin C promotes tumorigenesis in bladder cancer by activating the Wnt/beta-catenin signalling pathway. Front Biosci. 2024;29:327.
Zhang GP, Yue X, Li SQ. Cathepsin C interacts with TNF-alpha/p38 MAPK signaling pathway to promote proliferation and metastasis in hepatocellular carcinoma. Cancer Res Treat. 2020;52:10–23.
Xiao Y, Cong M, Li J, He D, Wu Q, Tian P, et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell. 2021;39:423–37 e7.
Eom T, Zhang C, Wang H, Lay K, Fak J, Noebels JL, et al. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife. 2013;2:e00178.
Nikom D, Zheng S. Alternative splicing in neurodegenerative disease and the promise of RNA therapies. Nat Rev Neurosci. 2023;24:457–73.
Mehta PR, Brown AL, Ward ME, Fratta P. The era of cryptic exons: implications for ALS-FTD. Mol Neurodegener. 2023;18:16.
Wilkins OG, Chien M, Wlaschin JJ, Barattucci S, Harley P, Mattedi F, et al. Creation of de novo cryptic splicing for ALS and FTD precision medicine. Science. 2024;386:61–9.
Liao H, Wang S, Wang X, Dai DZ, Zhang Y, Zhu C, et al. Imaging-assisted antisense oligonucleotide delivery for tumor-targeted gene therapy. Chem Biomed Imaging. 2024;2:313–30.
Cao M, Yan J, Ding Y, Zhang Y, Sun Y, Jiang G, et al. The potential impact of RNA splicing abnormalities on immune regulation in endometrial cancer. Cell Death Dis. 2025;16:148.
Acknowledgements
This work was partially supported by the China Health Promotion Foundation. The authors sincerely thank Xinyue Ma and Changjian Qi from our lab for their assistance with paraffin section preparation and the establishment of the PDX model. We thank American Journal Experts (AJE) for English language editing. The authors would also like to express their sincere gratitude to Ms. Qi Qi for her valuable suggestions on language editing.
Author information
Authors and Affiliations
Contributions
Conception and design: Yingwei Li. Methodology: Yingwei Li. Acquisition of data: Zhongshao Chen, Yanling Liu, and Yuehan Gao. Analysis and interpretation of data: Zhongshao Chen. Technical, or material support: Qianqian Gao, Yingying Pu, Feng Gao, and Ning Yang. Study supervision: Peng Li. Writing, review, and/or revision of the manuscript: Yingwei Li and Zhongshao Chen. Final approval: all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
Informed consent was obtained from all participants. Ethical approval was granted by the Ethics Committee of Qilu Hospital, Shandong University (KYLL-202210-055-1). The animal study was approved by the Animal Care and Use Committee of Shandong University (25038). All methods were performed in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by: Dr Barak Rotblat.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Chen, Z., Liu, Y. et al. SNRPD2-mediated regulation of DDX39B splicing promotes endometrial cancer progression by suppressing the activation of CTSC cryptic exons. Cell Death Dis (2026). https://doi.org/10.1038/s41419-026-08489-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-026-08489-4