Abstract
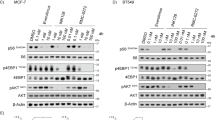
Breast cancer (BC) is the most prevalent malignancy in women, with hormone receptor-positive, HER2-negative (HR+/HER2−) tumors representing ~70% of cases. While CDK4/6 inhibitors (CDK4/6i) combined with endocrine therapy have transformed treatment for metastatic HR+/HER2− BC, acquired resistance remains a major obstacle. Using HR+/HER2− BC models with acquired resistance to the CDK4/6 inhibitors Palbociclib or Ribociclib, we uncovered a metabolic vulnerability in highly resistant clones, mediated by mTORC1 hyperactivation and autophagy suppression. Gene expression profiling revealed enrichment of glycolysis and mTORC1 pathways in CDK4/6i-resistant cells, which manifested as heightened sensitivity to the metabolic inhibitors Metformin and Dichloroacetate (DCA). Mechanistically, mTORC1 overactivation impaired autophagy via ULK1-Ser757 phosphorylation, as confirmed by LC3 flux assays, leaving resistant cells unable to adapt to energy stress. Treatment with metabolic drugs triggered AMPK activation, ACC inhibition, and PARP cleavage, culminating in apoptosis. Clinically, immunohistochemical analysis of a BC cohort revealed a significant correlation between mTORC1 activity (p4E-BP1T37/46) and autophagy suppression (p62 accumulation), supporting the translational relevance of this axis. Our findings propose mTORC1-mediated autophagy defects as a biomarker for metabolic vulnerability in CDK4/6i-resistant BC, offering a rationale for targeting these tumors with metabolic therapies to overcome resistance.
Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary figures). The following source western blot data are provided with this paper: uncropped images for for Figs. 3d, 4c and 4d. The GSEA data generated and analyzed for this paper are available at BioStudies with accession code E-MTAB-15265.
References
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63.
Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol. 2022;95:20211033.
Gremke N, Wagner U, Kalder M, Kostev K. Changes in the incidence of early-onset breast cancer in Germany between 2010 and 2022. Breast Cancer Res Treat. 2023;202:167–172.
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74.
Giaquinto AN, Sung H, Newman LA, Freedman RA, Smith RA, Star J, et al. Breast cancer statistics 2024. CA Cancer J Clin. 2024;74:477–95.
Ding L, Cao J, Lin W, Chen H, Xiong X, Ao H, et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci. 2020;21:1960.
Huppert LA, Gumusay O, Idossa D, Rugo HS. Systemic therapy for hormone receptor-positive/human epidermal growth factor receptor 2-negative early stage and metastatic breast cancer. CA Cancer J Clin. 2023;73:480–515.
Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Hart L, et al. Overall survival with Ribociclib plus Letrozole in advanced breast cancer. N Engl J Med. 2022;386:942–50.
Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer. Ann Oncol. 2018;29:1541–7.
Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35.
Goetz MP, Toi M, Campone M, Sohn J, Paluch-Shimon S, Huober J, et al. MONARCH 3: Abemaciclib as initial therapy for advanced breast cancer. J Clin Oncol. 2017;35:3638–46.
Dickler MN, Tolaney SM, Rugo HS, Cortés J, Diéras V, Patt D, et al. MONARCH 1, a phase II study of Abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR(+)/HER2(-) metastatic breast cancer. Clin Cancer Res. 2017;23:5218–24.
Slamon DJ, Neven P, Chia S, Fasching PA, De Laurentiis M, Im SA, et al. Phase III randomized study of ribociclib and fulvestrant in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: MONALEESA-3. J Clin Oncol. 2018;36:2465–72.
Álvarez-Fernández M, Malumbres M. Mechanisms of Sensitivity and Resistance to CDK4/6 Inhibition. Cancer Cell. 2020;37:514–29.
Gremke N, Rodepeter FR, Teply-Szymanski J, Griewing S, Boekhoff J, Stroh A, et al. NGS-guided precision oncology in breast cancer and gynecological tumors-a retrospective molecular tumor board analysis. Cancers. 2024;16:1561.
Pandey K, An HJ, Kim SK, Lee SA, Kim S, Lim SM, et al. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: a review. Int J Cancer. 2019;145:1179–88.
Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–76.
Deleyto-Seldas N, Efeyan A. The mTOR-autophagy axis and the control of metabolism. Front Cell Dev Biol. 2021;9:655731.
Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41.
Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64.
Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41.
Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. 2017;25:1037–43.
Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47.
Polo P, Gremke N, Stiewe T, Wanzel M. Robustness of the autophagy pathway to somatic copy number losses. Cells. 2022;11:1762.
Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–84.
Gremke N, Besong I, Stroh A, von Wichert L, Witt M, Elmshäuser S, et al. Targeting PI3K inhibitor resistance in breast cancer with metabolic drugs. Signal Transduction and Targeted Therapy. 2025;10:92.
Gremke N, Polo P, Dort A, Schneikert J, Elmshäuser S, Brehm C, et al. mTOR-mediated cancer drug resistance suppresses autophagy and generates a druggable metabolic vulnerability. Nat Commun. 2020;11:4684.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–80.
O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13:417–30.
Foretz M, Guigas B, Viollet B. Metformin: update on mechanisms of action and repurposing potential. Nat Rev Endocrinol. 2023;19:460–76.
Pang J, Li H, Sheng Y. CDK4/6 inhibitor resistance: a bibliometric analysis. Front Oncol. 2022;12:917707.
Wander SA, Cohen O, Gong X, Johnson GN, Buendia-Buendia JE, Lloyd MR, et al. The genomic landscape of intrinsic and acquired resistance to cyclin-dependent kinase 4/6 inhibitors in patients with hormone receptor-positive metastatic breast cancer. Cancer Discov. 2020;10:1174–93.
Mo H, Liu X, Xue Y, Chen H, Guo S, Li Z, et al. S6K1 amplification confers innate resistance to CDK4/6 inhibitors through activating c-Myc pathway in patients with estrogen receptor-positive breast cancer. Mol Cancer. 2022;21:171.
Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020;9:2308.
Schwinn MK, Machleidt T, Zimmerman K, Eggers CT, Dixon AS, Hurst R, et al. CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem Biol. 2018;13:467–74.
Schläfli AM, Berezowska S, Adams O, Langer R, Tschan MP. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem. 2015;59:2481.
Huang CI, Wang CC, Tai TS, Hwang TZ, Yang CC, Hsu CM, et al. eIF4E and 4EBP1 are prognostic markers of head and neck squamous cell carcinoma recurrence after definitive surgery and adjuvant radiotherapy. PLoS One. 2019;14:e0225537.
Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–40.
Zhou FH, Downton T, Freelander A, Hurwitz J, Caldon CE, Lim E. CDK4/6 inhibitor resistance in estrogen receptor positive breast cancer, a 2023 perspective. Front Cell Dev Biol. 2023;11:1148792.
Presti D, Quaquarini E The PI3K/AKT/mTOR and CDK4/6 pathways in endocrine resistant HR+/HER2- metastatic breast cancer: biological mechanisms and new treatments. Cancers. 2019;11:1242.
Romero-Pozuelo J, Figlia G, Kaya O, Martin-Villalba A, Teleman AA. Cdk4 and Cdk6 Couple the cell-cycle machinery to cell growth via mTORC1. Cell Rep. 2020;31:107504.
Zacharek SJ, Xiong Y, Shumway SD. Negative regulation of TSC1-TSC2 by mammalian D-type cyclins. Cancer Res. 2005;65:11354–60.
Abu-Khalaf MM, Alex Hodge K, Hatzis C, Baldelli E, El Gazzah E, Valdes F, et al. AKT/mTOR signaling modulates resistance to endocrine therapy and CDK4/6 inhibition in metastatic breast cancers. NPJ Precis Oncol. 2023;7:18.
Rodriguez MJ, Perrone MC, Riggio M, Palafox M, Salinas V, Elia A, et al. Targeting mTOR to overcome resistance to hormone and CDK4/6 inhibitors in ER-positive breast cancer models. Sci Rep. 2023;13:2710.
Cai Z, Wang J, Li Y, Shi Q, Jin L, Li S, et al. Overexpressed Cyclin D1 and CDK4 proteins are responsible for the resistance to CDK4/6 inhibitor in breast cancer that can be reversed by PI3K/mTOR inhibitors. Sci China Life Sci. 2023;66:94–109.
O’Brien NA, McDermott MSJ, Conklin D, Luo T, Ayala R, Salgar S, et al. Targeting activated PI3K/mTOR signaling overcomes acquired resistance to CDK4/6-based therapies in preclinical models of hormone receptor-positive breast cancer. Breast Cancer Res. 2020;22:89.
Huber K, Mestres-Arenas A, Fajas L, Leal-Esteban LC. The multifaceted role of cell cycle regulators in the coordination of growth and metabolism. FEBS J. 2021;288:3813–33.
Lee Y, Dominy JE, Choi YJ, Jurczak M, Tolliday N, Camporez JP, et al. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510:547–51.
Qie S, Yoshida A, Parnham S, Oleinik N, Beeson GC, Beeson CC, et al. Targeting glutamine-addiction and overcoming CDK4/6 inhibitor resistance in human esophageal squamous cell carcinoma. Nat Commun. 2019;10:1296.
Hu Q, Peng J, Jiang L, Li W, Su Q, Zhang J, et al. Metformin as a senostatic drug enhances the anticancer efficacy of CDK4/6 inhibitor in head and neck squamous cell carcinoma. Cell Death Dis. 2020;11:925.
Li N, Sun YJ, Huang LY, Li RR, Zhang JS, Qiu AH, et al. Fasting-mimicking diet potentiates anti-tumor effects of CDK4/6 inhibitors against breast cancer by suppressing NRAS- and IGF1-mediated mTORC1 signaling. Drug Resist Updat. 2025;78:101161.
Ma Y, Zhu Q, Liang J, Li Y, Li M, Zhang Y, et al. A CRISPR knockout negative screen reveals synergy between CDKs inhibitor and metformin in the treatment of human cancer in vitro and in vivo. Signal Transduct Target Ther. 2020;5:152.
Vijayaraghavan S, Karakas C, Doostan I, Chen X, Bui T, Yi M, et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat Commun. 2017;8:15916.
Gong C, Lin Q, Qin T, Zeng Y, Xu F, Yang Y, et al. Targeting autophagy plus high-dose CDK4/6 inhibitors in advanced HR+HER2- breast cancer: a phase 1b/2 trial. Med. 2024;6:100559.
Tang H, Sebti S, Titone R, Zhou Y, Isidoro C, Ross TS, et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine. 2015;2:255–63.
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222.
Arafeh R, Shibue T, Dempster JM, Hahn WC, Vazquez F. The present and future of the Cancer Dependency Map. Nat Rev Cancer. 2025;25:59–73.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–32.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Acknowledgements
We extend our gratitude to Marina Föth, Sigrid Bischofsberger, and Viktoria Wischmann for their experimental contributions and technical assistance. We also thank the members of our lab for their valuable discussions and advice.
Funding
NG was supported by the Clinician Scientist Program (SUCCESS-Program) of Philipps University Marburg and the UniversityHospital of Giessen and Marburg (UKGM), as well as the Clinical Trialist Program (SUCCESS meets MSNZ) of the Department ofMedicine. NG received research funding from the Deutsche Forschungsgemeinschaft (DFG) (GRK 2573/2 – 2024), the UniversityMedical Center Giessen and Marburg (UKGM) (Grant 3/2022 MR to NG), the von Behring-Röntgen Foundation (Grant 70_0027 toNG), the PE Kempkes Foundation (Grant 01/2021 to NG), and the Medical Foundation (Grant 04/2021 to NG). Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
NG and TS jointly planned and supervised the study. NG generated CDK4/6 inhibitor-resistant clones. LvW performed most experiments with contributions from NG, AS and MWitt. NG, LvW and JT-S prepared and analyzed samples using HTG EdgeSeq. M.M. performed GSEA analyses. BMP and CD provided breast cancer tissue microarrays and clinical data. LvW and A-SL performed immunohistochemistry and statistical analysis of the tissue microarrays. TW, UW, TS and NG provided resources. M Wanzel, SG, TW and UW provided essential intellectual and conceptual input. LvW, NG and TS analyzed data, performed statistics and wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
CD received personal fees from Novartis, Roche, MSD Oncology, Daiichi Sankyo, AstraZeneca, Molecular Health, and Merck, all outside the submitted work. CD is cofounder of Sividon Diagnostics. In addition, CD has a patent on VMScope digital pathology software with royalties paid; a patent WO2020109570A1—cancer immunotherapy pending; and patents WO2015114146A1 and WO2010076322A1—therapy response issued. PJ reports research grant and travel expenses from GILEAD Sciences GmbH out-side the submitted work. NH declares to be GBG Forschungs GmbH employee. GBG Forschungs GmbH reports financial funding from AstraZeneca and Myriad during the conduct of the study; received funding for research grants from Abbvie, Amgen, AstraZeneca, BMS, Daiichi-Sankyo, Gilead, Molecular Health, Stemline Menarini, Celgene/BMS, Novartis, Pfizer and Roche (paid to the institution); GBG Forschungs GmbH has licensing fees from VMscope GmbH. In addition, GBG Forschungs GmbH has a patent EP21152186.9 pending, a patent EP19808852.8 pending, and a patent EP14153692.0 pending. The other authors declare no conflicts of interest.
Ethics approval and consent to participate
Immunohistochemical staining and evaluation of breast cancer patient tissue samples were approved by the Ethics Committee of the Charité (Ethics Opinion No. EA1/139/05) and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients for the use of their tissue samples in this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Dr Satoshi Inoue
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
von Wichert, L., Stroh, A., Witt, M. et al. mTOR-driven autophagy suppression defines metabolic vulnerability in CDK4/6 inhibitor-resistant HR+/HER2− breast cancer. Cell Death Dis (2026). https://doi.org/10.1038/s41419-026-08496-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-026-08496-5