Abstract
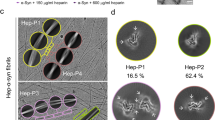
α-Synuclein (α-syn) amyloid fibrils are the major component of Lewy bodies, which are the pathological hallmark of Parkinson’s disease (PD) and other synucleinopathies. High-resolution structure of α-syn fibril is important for understanding its assembly and pathological mechanism. Here, we determined a fibril structure of full-length α-syn (1–140) at the resolution of 3.07 Å by cryo-electron microscopy (cryo-EM). The fibrils are cytotoxic, and transmissible to induce endogenous α-syn aggregation in primary neurons. Based on the reconstructed cryo-EM density map, we were able to unambiguously build the fibril structure comprising residues 37–99. The α-syn amyloid fibril structure shows two protofilaments intertwining along an approximate 21 screw axis into a left-handed helix. Each protofilament features a Greek key-like topology. Remarkably, five out of the six early-onset PD familial mutations are located at the dimer interface of the fibril (H50Q, G51D, and A53T/E) or involved in the stabilization of the protofilament (E46K). Furthermore, these PD mutations lead to the formation of fibrils with polymorphic structures distinct from that of the wild-type. Our study provides molecular insight into the fibrillar assembly of α-syn at the atomic level and sheds light on the molecular pathogenesis caused by familial PD mutations of α-syn.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Goedert, M., Clavaguera, F. & Tolnay, M. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci. 33, 317–325 (2010).
Jucker, M. & Walker, L. C. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 (2013).
Knowles, T. P., Vendruscolo, M. & Dobson, C. M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014).
Tycko, R. Physical and structural basis for polymorphism in amyloid fibrils. Protein Sci. 23, 1528–1539 (2014).
Riek, R. & Eisenberg, D. S. The activities of amyloids from a structural perspective. Nature 539, 227–235 (2016).
Qiang, W., Yau, W. M., Lu, J. X., Collinge, J. & Tycko, R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 541, 217–221 (2017).
Rodriguez, J. A. et al. Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015).
Gremer, L. et al. Fibril structure of amyloid-β(1-42) by cryo-electron microscopy. Science 358, 116–119 (2017).
Fitzpatrick, A. W. P. et al. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017).
Spillantini, M. G. et al. alpha-Synuclein in Lewy bodies. Nature 388, 839–840 (1997).
Goedert, M. & Spillantini, M. G. Lewy body diseases and multiple system atrophy as alpha-synucleinopathies. Mol. Psychiatry 3, 462–465 (1998).
Goedert, M., Spillantini, M. G., Del Tredici, K. & Braak, H. 100 years of Lewy pathology. Nat. Rev. Neurol. 9, 13–24 (2013).
Moore, D. J., West, A. B., Dawson, V. L. & Dawson, T. M. Molecular pathophysiology of Parkinson’s disease. Annu. Rev. Neurosci. 28, 57–87 (2005).
Chartier-Harlin, M. C. et al. α-Synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364, 1167–1169 (2004).
Singleton, A. B. et al. α-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841 (2003).
Krüger, R. et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 18 106–108 (1998).
Zarranz, JuanJ. et al. The new mutation, E46K, of α-synuclein causes parkinson and Lewy body dementia. Ann. Neuro. 55, 164–173 (2004).
Appel-Cresswell, S. et al. Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov. Disord. 28, 811–813 (2013).
Lesage, S. et al. G51D alpha-synuclein mutation causes a novel parkinsonian-pyramidal syndrome. Ann. Neuro. 73, 459–471 (2013).
Pasanen, P. et al. Novel α-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol. Aging 35, 2180.e2181–2185 (2014).
Polymeropoulos, M. H. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997).
Goedert, M., Jakes, R. & Spillantini, M. G. The synucleinopathies: twenty years on. J. Park. Dis. 7, S53–S71 (2017).
Peelaerts, W. et al. alpha-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344 (2015).
Guo, J. L. et al. Distinct α-synuclein strains differentially promote tau inclusions in neurons. Cell 154, 103–117 (2013).
Peng, C. et al. Cellular milieu imparts distinct pathological alpha-synuclein strains in alpha-synucleinopathies. Nature 557, 558–563 (2018).
Theillet, F. X. et al. Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature 530, 45–50 (2016).
Tuttle, M. D. et al. Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat. Struct. Mol. Bio. 23, 409–415 (2016).
Choi, W. et al. Mutation E46K increases phospholipid binding and assembly into filaments of human alpha-synuclein. FEBS Lett. 576, 363–368 (2004).
Khalaf, O. et al. The H50Q mutation enhances α-synuclein aggregation, secretion, and toxicity. J. Biol. Chem. 289, 21856–21876 (2014).
Jo, E., Fuller, N., Rand, R. P., St George-Hyslop, P. & Fraser, P. E. Defective membrane interactions of familial Parkinson’s disease mutant A30P alpha-synuclein. J. Mol. Biol. 315, 799–807 (2002).
Flagmeier, P. et al. Mutations associated with familial Parkinson’s disease alter the initiation and amplification steps of alpha-synuclein aggregation. Proc. Natl Acad. Sci. USA 113, 10328–10333 (2016).
Reynolds, N. P. et al. Mechanism of membrane interaction and disruption by alpha-synuclein. J. Am. Chem. Soc. 133, 19366–19375 (2011).
Fares, M. B. et al. The novel Parkinson’s disease linked mutation G51D attenuates in vitro aggregation and membrane binding of alpha-synuclein, and enhances its secretion and nuclear localization in cells. Hum. Mol. Genet. 23, 4491–4509 (2014).
Lazaro, D. F. et al. Systematic comparison of the effects of alpha-synuclein mutations on its oligomerization and aggregation. PLoS ONE 10, e1004741 (2014).
Guerrero-Ferreira, R. et al. Cryo-EM structure of alpha-synuclein fibrils. eLife 7, e36402 (2018).
Johnson, M., Coulton, A. T., Geeves, M. A. & Mulvihill, D. P. Targeted amino-terminal acetylation of recombinant proteins in E. coli. PLoS ONE 5, e15801 (2010).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63 (2014).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V., Headd, J. J., Terwilliger, T. C. & Adams, P. D. New tool: phenix.real_space_refine. Comp. Cryst. Newsl. 4, 43–44 (2013).
Acknowledgements
This work was supported by the Major State Basic Research Development Program (2016YFA0501902 to C.L., X.L. and 2016YFA0501102 to X.L.), the National Natural Science Foundation (NSF) of China (31470748 to C.L. and 31570730 and 31722015 to X.L.), the Chinese Academy of Sciences (to C.L.), The “1000 Talents Plan” of China to C.L. and X.L., the Advanced Innovation Center for Structural Biology (to X.L.), Tsinghua-Peking Joint Center for Life Sciences (to X.L.), Shanghai Pujiang Program (to D.L.). We acknowledge Tsinghua University Branch of China National Center for Protein Sciences Beijing for providing facility supports in cryo-EM.
Author information
Authors and Affiliations
Contributions
X.L., C.L., D.L., Y.L. and C.Z. designed the project. Y.L. and F.L. performed the cryo-electron microscopy experiments. Y.L. and Z.Luo built and refined the structure model. C.Z., X.G. and X.Z. performed the biochemical and cellular assays. Z.Liu performed the AFM experiments. Y.L., C.Z., D.L., X.L. and C.L. analyzed the data and contributed to manuscript discussion and editing. D.L. and X.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Li, Y., Zhao, C., Luo, F. et al. Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res 28, 897–903 (2018). https://doi.org/10.1038/s41422-018-0075-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-018-0075-x
Keywords
This article is cited by
-
C-terminus-dependent detection of lysosomal alpha-synuclein in nigral Parkinson’s disease human brain neurons
Molecular Neurodegeneration (2025)
-
Surface wetting is a key determinant of α-synuclein condensate maturation
Communications Chemistry (2025)
-
Design of Ig-like binders targeting α-synuclein fibril for mitigating its pathological activities
Nature Communications (2025)
-
Defining essential charged residues in fibril formation of a lysosomal derived N-terminal α-synuclein truncation
Nature Communications (2025)
-
Aspirin inhibits proteasomal degradation and promotes α-synuclein aggregate clearance through K63 ubiquitination
Nature Communications (2025)