Abstract
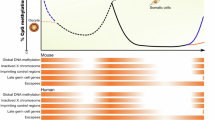
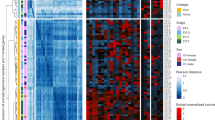
Extensive and accurate chromatin remodeling is essential during primordial germ cell (PGC) development for the perpetuation of genetic information across generations. Here, we report that distal cis-regulatory elements (CREs) marked by DNase I-hypersensitive sites (DHSs) show temporally restricted activities during mouse and human PGC development. Using DHS maps as proxy, we accurately locate the genome-wide binding sites of pluripotency transcription factors in mouse PGCs. Unexpectedly, we found that mouse female meiotic recombination hotspots can be captured by DHSs, and for the first time, we identified 12,211 recombination hotspots in mouse female PGCs. In contrast to that of meiotic female PGCs, the chromatin of mitotic-arrested male PGCs is permissive through nuclear transcription factor Y (NFY) binding in the distal regulatory regions. Furthermore, we examined the evolutionary pressure on PGC CREs, and comparative genomic analysis revealed that mouse and human PGC CREs are evolutionarily conserved and show strong conservation across the vertebrate tree outside the mammals. Therefore, our results reveal unique, temporally accessible chromatin configurations during mouse and human PGC development.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Tang, W. W., Kobayashi, T., Irie, N., Dietmann, S. & Surani, M. A. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 17, 585–600 (2016).
Saitou, M. & Yamaji, M. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 4, pii: a008375 (2012).
Spiller, C., Wilhelm, D. & Koopman, P. Cell cycle analysis of fetal germ cells during sex differentiation in mice. Biol. Cell 101, 587–598 (2009).
Western, P. S., Miles, D. C., van den Bergen, J. A., Burton, M. & Sinclair, A. H. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 26, 339–347 (2008).
Guo, F. et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161, 1437–1452 (2015).
Li, L. et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell. Stem. Cell. 20, 858–873 (2017).
Seisenberger, S. et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862 (2012).
Kobayashi, H. et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 23, 616–627 (2013).
Lu, F. et al. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165, 1375–1388 (2016).
Nord, A. S. et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531 (2013).
Leitch, H. G. & Smith, A. The mammalian germline as a pluripotency cycle. Development 140, 2495–2501 (2013).
Kurimoto, K. et al. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell. Stem. Cell. 16, 517–532 (2015).
Hajkova, P. et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881 (2008).
Long, H. K., Prescott, S. L. & Wysocka, J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016).
Villar, D. et al. Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015).
Lesch, B. J., Dokshin, G. A., Young, R. A., McCarrey, J. R. & Page, D. C. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc. Natl Acad. Sci. USA 110, 16061–16066 (2013).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Vierstra, J. et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346, 1007–1012 (2014).
Soucie, E. L. et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science 351, aad5510 (2016).
Lara-Astiaso, D. et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Neph, S. et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90 (2012).
Runyan, C. et al. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development 133, 4861–4869 (2006).
Hesselberth, J. R. et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 (2009).
Buecker, C. et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell. Stem. Cell. 14, 838–853 (2014).
Li, M. & Belmonte, J. C. Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 18, 180–191 (2017).
He, H. H. et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods 11, 73–78 (2014).
Kojima, Y. et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem. Cell 14, 107–120 (2014).
Yamaji, M. et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40, 1016–1022 (2008).
Brick, K., Smagulova, F., Khil, P., Camerini-Otero, R. D. & Petukhova, G. V. Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012).
Khil, P. P., Smagulova, F., Brick, K. M., Camerini-Otero, R. D. & Petukhova, G. V. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 22, 957–965 (2012).
Smagulova, F. et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375–378 (2011).
Baudat, F. et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 (2010).
Davies, B. et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530, 171–176 (2016).
Kelmenson, P. M. et al. torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics 169, 833–841 (2005).
Walker, M. et al. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin 8, 31 (2015).
Tsai, I. J., Burt, A. & Koufopanou, V. Conservation of recombination hotspots in yeast. Proc. Natl Acad. Sci. USA 107, 7847–7852 (2010).
Pratto, F. et al. DNA recombination. Recombination initiation maps of individual human genomes. Science 346, 1256442 (2014).
Nardone, V., Chaves-Sanjuan, A. & Nardini, M. Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta 1860, 571–580 (2017).
Nardini, M. et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152, 132–143 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Oldfield, A. J. et al. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55, 708–722 (2014).
Bungartz, G., Land, H., Scadden, D. T. & Emerson, S. G. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood 119, 1380–1389 (2012).
Hammoud, S. S. et al. Transcription and imprinting dynamics in developing postnatal male germline stem cells. Genes Dev. 29, 2312–2324 (2015).
Kanatsu-Shinohara, M. & Shinohara, T. Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell. Dev. Biol. 29, 163–187 (2013).
Meng, X. et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493 (2000).
He, Z. et al. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells 26, 266–278 (2008).
Whittle, C. A. & Extavour, C. G. Causes and evolutionary consequences of primordial germ-cell specification mode in metazoans. Proc. Natl Acad. Sci. USA 114, 5784–5791 (2017).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Blow, M. J. et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42, 806–810 (2010).
Baudat, F., Imai, Y. & de Massy, B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 14, 794–806 (2013).
Cotney, J. et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154, 185–196 (2013).
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, H. et al. The Sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
John, S. et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 (2011).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Hedges, S. B. The origin and evolution of model organisms. Nat. Rev. Genet. 3, 838–849 (2002).
Murphy, W. J., Pringle, T. H., Crider, T. A., Springer, M. S. & Miller, W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 17, 413–421 (2007).
Bininda-Emonds, O. R. et al. The delayed rise of present-day mammals. Nature 446, 507–512 (2007).
Acknowledgements
We are grateful to our colleagues in the laboratory for their assistance with the experiments and in the preparation of this manuscript. This work was primarily supported by the National Key R&D Program of China (2016YFA0100400) and the National Natural Science Foundation of China (31721003). This work was also supported by the Ministry of Science and Technology of China (2017YFA0102602 and 2015CB964800), the National Natural Science Foundation of China (81630035, 31771419, 31871446, 31871448 and 81501250), the Shanghai Rising-Star Program (17QA1402700), the Shanghai Chenguang Program (16CG17 and 16CG19), the Shanghai Municipal Medical and Health Discipline Construction Projects (2017ZZ02015), and the Shanghai Municipal Commission of Health and Family Planning (201540364 and 20164037).
Author contributions
J.L. and S.G. designed the experiments; J.L. conducted the experiments with assistance from J.C., W.L., X.L., B.W., J.Y., L.W., M.W., L.G. and H.W.; J.L., S.S. and C.J. analyzed the data with assistance from Q.Z. and X.C.; J.L., S.S., C.J. and S.G. wrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, J., Shen, S., Chen, J. et al. Accurate annotation of accessible chromatin in mouse and human primordial germ cells. Cell Res 28, 1077–1089 (2018). https://doi.org/10.1038/s41422-018-0096-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-018-0096-5
Keywords
This article is cited by
-
Binding of NF-Y to transposable elements in mouse and human cells
Mobile DNA (2025)
-
Intrauterine hyperglycemia impairs mouse primordial germ cell development and fertility by sex-specific epigenetic reprogramming interference
Cell Discovery (2025)
-
Single-cell long-read Hi-C, scNanoHi-C2, details 3D genome reorganization in embryonic-stage germ cells
Nature Structural & Molecular Biology (2025)
-
The role(s) of NF-Y in development and differentiation
Cell Death & Differentiation (2025)
-
MAX controls meiotic entry in sexually undifferentiated germ cells
Scientific Reports (2024)