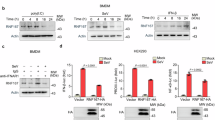
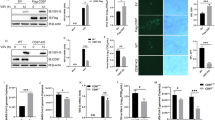
Abstract
Autophagy is a conserved process that delivers cytosolic substances to the lysosome for degradation, but its direct role in the regulation of antiviral innate immunity remains poorly understood. Here, through high-throughput screening, we discovered that CCDC50 functions as a previously unknown autophagy receptor that negatively regulates the type I interferon (IFN) signaling pathway initiated by RIG-I-like receptors (RLRs), the sensors for RNA viruses. The expression of CCDC50 is enhanced by viral infection, and CCDC50 specifically recognizes K63-polyubiquitinated RLRs, thus delivering the activated RIG-I/MDA5 for autophagic degradation. The association of CCDC50 with phagophore membrane protein LC3 is confirmed by crystal structure analysis. In contrast to other known autophagic cargo receptors that associate with either the LIR-docking site (LDS) or the UIM-docking site (UDS) of LC3, CCDC50 can bind to both LDS and UDS, representing a new type of cargo receptor. In mouse models with RNA virus infection, CCDC50 deficiency reduces the autophagic degradation of RIG-I/MDA5 and promotes type I IFN responses, resulting in enhanced viral resistance and improved survival rates. These results reveal a new link between autophagy and antiviral innate immune responses and provide additional insights into the regulatory mechanisms of RLR-mediated antiviral signaling.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
X-ray crystallography data (PDB ID: 6LAN) and deep sequencing data (GEO: GSE143467) are deposited in the PDB and GEO databases, respectively. All of the other data supporting this research are available from the corresponding author upon request.
References
Chan, Y. K. & Gack, M. U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 14, 360–373 (2016).
Tan, X., Sun, L., Chen, J. & Chen, Z. J. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 72, 447–478 (2018).
Bruns, A. M. & Horvath, C. M. Antiviral RNA recognition and assembly by RLR family innate immune sensors. Cytokine Growth Factor Rev. 25, 507–512 (2014).
Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF3. Cell 122, 669–682 (2005).
Kuka, M., De Giovanni, M. & Iannacone, M. The role of type I interferons in CD4(+) T cell differentiation. Immunol. Lett. 215, 19–23 (2019).
McNab, F., Mayer-Barber, K., Sher, A., Wack, A. & O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015).
Lee, H. K., Lund, J. M., Ramanathan, B., Mizushima, N. & Iwasaki, A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 315, 1398–1401 (2007).
Jounai, N. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA. 104, 14050–14055 (2007).
Lei, Y. et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 36, 933–946 (2012).
Du, Y. et al. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 37, 351–366 (2018).
Chen, M. et al. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to Promote innate immune responses. Mol. Cell 64, 105–119 (2016).
Prabakaran, T. et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 37, e97858 (2018).
Jin, S. et al. Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol. Cell 68, 308–322 (2017).
Vazza, G., Picelli, S., Bozzato, A. & Mostacciuolo, M. L. Identification and characterization of C3orf6, a new conserved human gene mapping to chromosome 3q28. Gene 314, 113–120 (2003).
Tashiro, K. et al. Suppression of the ligand-mediated down-regulation of epidermal growth factor receptor by Ymer, a novel tyrosine-phosphorylated and ubiquitinated protein. J. Biol. Chem. 281, 24612–24622 (2006).
Tsukiyama, T. et al. Ymer acts as a multifunctional regulator in nuclear factor-kappaB and Fas signaling pathways. Mol. Med. 18, 587–597 (2012).
Farfsing, A. et al. Gene knockdown studies revealed CCDC50 as a candidate gene in mantle cell lymphoma and chronic lymphocytic leukemia. Leukemia 23, 2018–2026 (2009).
Hong, W. et al. A CCDC50 splice variant is modulated by SRSF3 and promotes hepatocellular carcinoma via the Ras signaling pathway. Hepatology 69, 179–195 (2018).
Ferri, F. et al. TRIM33 switches off Ifnb1 gene transcription during the late phase of macrophage activation. Nat. Commun. 6, 8900 (2015).
Liu, S. et al. Lck/Hck/Fgr-mediated tyrosine phosphorylation negatively regulates TBK1 to restrain innate antiviral responses. Cell Host Microbe 21, 754–768 (2017).
Deng, W. et al. The novel secretory protein CGREF1 inhibits the activation of AP-1 transcriptional activity and cell proliferation. Int. J. Biochem. Cell Biol. 65, 32–39 (2015).
Kaneda, M. M. et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature 539, 437–442 (2016).
Tan, J. M. et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 17, 431–439 (2008).
Penengo, L. et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124, 1183–1195 (2006).
Johansen, T. & Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011).
Marshall, R. S., Hua, Z., Mali, S., McLoughlin, F. & Vierstra, R. D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766–781 (2019).
Suzuki, H. et al. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure 22, 47–58 (2014).
Birgisdottir, A. B., Lamark, T. & Johansen, T. The LIR motif—crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 (2013).
Consortium, G. T. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Choi, Y., Bowman, J. W. & Jung, J. U. Autophagy during viral infection—a double-edged sword. Nat. Rev. Microbiol. 16, 341–354 (2018).
Levine, B., Mizushima, N. & Virgin, H. W. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011).
Deretic, V., Saitoh, T. & Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737 (2013).
Richetta, C. et al. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 9, e1003599 (2013).
Watson, R. O. et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17, 811–819 (2015).
Lamark, T., Svenning, S. & Johansen, T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 61, 609–624 (2017).
Ablasser, A. & Hur, S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 21, 17–29 (2019).
Gack, M. U. et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007).
Cadena, C. et al. Ubiquitin-dependent and -independent roles of E3 ligase RIPLET in innate immunity. Cell 177, 1187–1200 (2019).
Jiang, X. et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36, 959–973 (2012).
Kirkin, V. et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009).
Shi, J. et al. NBR1 is dispensable for PARK2-mediated mitophagy regardless of the presence or absence of SQSTM1. Cell Death Dis. 6, e1943 (2015).
Gonzalez-Navajas, J. M., Lee, J., David, M. & Raz, E. Immunomodulatory functions of type I interferons. Nat . Rev. Immunol. 12, 125–135 (2012).
Mann, D. L. The emerging role of innate immunity in the heart and vascular system: for whom the cell tolls. Circ. Res. 108, 1133–1145 (2011).
Hou, P. et al. A genome-wide CRISPR screen identifies genes critical for resistance to FLT3 inhibitor AC220. Cancer Res. 77, 4402–4413 (2017).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102, 15545–15550 (2005).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Hou, P. et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 5, 15577 (2015).
Li, S. et al. The tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat. Immunol. 17, 241–249 (2016).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta. Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta. Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta. Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Acknowledgements
We thank Dr. Tian Tian (Center of Applied Genomics, Children’s Hospital of Philadelphia) for assistance with the bioinformatic analysis and Dr. Bo Zhong (Wuhan University) for providing the Cre-ER mice and EMCV. We also thank Dr. Jun Cui (Sun Yat-sen University) for gifting p62- and BECN1-deficient HEK293T cells. The research is supported by the National Natural Science Foundation of China (#81620108020 to D.G. and #81801574 to P.H.) and the China postdoctoral science foundation (2018M633238 to P.H.). D.G. is also supported by Shenzhen Science and Technology Program (#KQTD20180411143323605), the Guangdong Zhujiang Talents Programme and the National Ten-thousand Talents Programme. P.H. is supported by the postdoctoral innovation talents supporting project (#BX20180390).
Author information
Authors and Affiliations
Contributions
D.G. conceived and supervised the research; D.G. and P.H. designed the experiments, analyzed the data and wrote the manuscript. P.H., K.Y., P.J., W.Z. and Y.L. performed the biochemical, cell biological and in vitro experiments; P.H., K.Y., P.J. and Z.L. performed the animal experiments and viral infections; L.L., J.L. and Y.L. completed the structural analysis. W.Z., J.P., S.C., S.G., C.L., H.P., J.W. and Y.L. helped with reagents and discussions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests. D.G. and P.H. are applying for a patent related to the functions of CCDC50 in therapy of viral infection and other diseases.
Supplementary information
Rights and permissions
About this article
Cite this article
Hou, P., Yang, K., Jia, P. et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res 31, 62–79 (2021). https://doi.org/10.1038/s41422-020-0362-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-020-0362-1
This article is cited by
-
Inhibition of STING-mediated antiviral innate immunity activation by CD97 via modulation of ER-phagy
Communications Biology (2025)
-
Exploring new drug treatment targets for immune related bone diseases using a multi omics joint analysis strategy
Scientific Reports (2025)
-
RNF167 mediates atypical ubiquitylation and degradation of RLRs via two distinct proteolytic pathways
Nature Communications (2025)
-
E3 Ubiquitin ligases Cbl-b and c-Cbl maintain the homeostasis of macrophages by regulating the M-CSF/M-CSFR signaling axis
Cell Death & Disease (2025)
-
SARS-CoV-2 nsp14 induces the m7G methylation and the sequestration of host RNAs in stress granules
Communications Biology (2025)