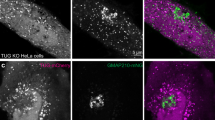
Abstract
Under stress, the endomembrane system undergoes reorganization to support autophagosome biogenesis, which is a central step in autophagy. How the endomembrane system remodels has been poorly understood. Here we identify a new type of membrane contact formed between the ER–Golgi intermediate compartment (ERGIC) and the ER-exit site (ERES) in the ER–Golgi system, which is essential for promoting autophagosome biogenesis induced by different stress stimuli. The ERGIC–ERES contact is established by the interaction between TMED9 and SEC12 which generates a short distance opposition (as close as 2–5 nm) between the two compartments. The tight membrane contact allows the ERES-located SEC12 to transactivate COPII assembly on the ERGIC. In addition, a portion of SEC12 also relocates to the ERGIC. Through both mechanisms, the ERGIC–ERES contact promotes formation of the ERGIC-derived COPII vesicle, a membrane precursor of the autophagosome. The ERGIC–ERES contact is physically and functionally different from the TFG-mediated ERGIC–ERES adjunction involved in secretory protein transport, and therefore defines a unique endomembrane structure generated upon stress conditions for autophagic membrane formation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Kawabata, T. & Yoshimori, T. Autophagosome biogenesis and human health. Cell Discov. 6, 33 (2020).
Morishita, H. & Mizushima, N. Diverse cellular roles of autophagy. Annu. Rev. Cell Dev. Biol. 35, 453–475 (2019).
Fujioka, Y. et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305 (2020).
Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458 (2020).
Nishimura, T. et al. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 14, 284–291 (2013).
Dooley, H. C. et al. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 55, 238–252 (2014).
Mizushima, N. et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116, 1679–1688 (2003).
Fujita, N. et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 (2008).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Slobodkin, M. R. & Elazar, Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 55, 51–64 (2013).
Ge, L., Baskaran, S., Schekman, R. & Hurley, J. H. The protein-vesicle network of autophagy. Curr. Opin. Cell Biol. 29, 18–24 (2014).
Schutter, M., Giavalisco, P., Brodesser, S. & Graef, M. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell 180, 135–149.e14 (2020).
Puri, C., Renna, M., Bento, C. F., Moreau, K. & Rubinsztein, D. C. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299 (2013).
Karanasios, E. et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7, 12420 (2016).
Osawa, T. et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281–288 (2019).
Valverde, D. P. et al. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787–1798 (2019).
Maeda, S., Otomo, C. & Otomo, T. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 8, e45777 (2019).
Nair, U. et al. SNARE proteins are required for macroautophagy. Cell 146, 290–302 (2011).
Moreau, K., Ravikumar, B., Renna, M., Puri, C. & Rubinsztein, D. C. Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317 (2011).
Nishimura, T. et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36, 1719–1735 (2017).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
Hayashi-Nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009).
Takahashi, Y. et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9, 2855 (2018).
Zhen, Y. et al. ESCRT-mediated phagophore sealing during mitophagy. Autophagy 16, 826–841 (2020).
Yu, L. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 (2010).
Ge, L., Melville, D., Zhang, M. & Schekman, R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2, e00947 (2013).
Ge, L. & Schekman, R. The ER-Golgi intermediate compartment feeds the phagophore membrane. Autophagy 10, 170–172 (2013).
Ge, L., Zhang, M. & Schekman, R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife 3, e04135 (2014).
Ge, L., Wilz, L. & Schekman, R. Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy 11, 2372–2374 (2015).
Gui, X. et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019).
Jeong, Y. T. et al. The ULK1-FBXW5-SEC23B nexus controls autophagy. Elife 7, e42253 (2018).
Scorrano, L. et al. Coming together to define membrane contact sites. Nat. Commun. 10, 1287 (2019).
Prinz, W. A., Toulmay, A. & Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 21, 7–24 (2020).
Cohen, S., Valm, A. M. & Lippincott-Schwartz, J. Interacting organelles. Curr. Opin. Cell Biol. 53, 84–91 (2018).
Xiao, J. et al. Cholesterol transport through the peroxisome-ER membrane contacts tethered by PI(4,5)P2 and extended synaptotagmins. Sci. China Life Sci. 62, 1117–1135 (2019).
Hamasaki, M. et al. Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013).
Nascimbeni, A. C. et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 36, 2018–2033 (2017).
Zhao, Y. G. et al. The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr. Biol. 28, 1234–1245 e1234 (2018).
Zhao, Y. G. et al. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell 67, 974–989.e976 (2017).
Johnson, A. et al. TFG clusters COPII-coated transport carriers and promotes early secretory pathway organization. EMBO J. 34, 811–827 (2015).
Hanna, M. G. T. et al. TFG facilitates outer coat disassembly on COPII transport carriers to promote tethering and fusion with ER-Golgi intermediate compartments. Proc. Natl. Acad. Sci. USA 114, E7707–E7716 (2017).
Witte, K. et al. TFG-1 function in protein secretion and oncogenesis. Nat. Cell Biol. 13, 550–558 (2011).
Noda, T. et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–480 (2000).
Morita, K. et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217, 3817–3828 (2018).
Shoemaker, C. J. et al. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 17, e2007044 (2019).
Ropolo, A. et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282, 37124–37133 (2007).
Moretti, F. et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 19, e45889 (2018).
Zhuang, X. et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 114, E426–E435 (2017).
Huang, D. et al. TMEM41B acts as an ER scramblase required for lipoprotein biogenesis and lipid homeostasis. Cell Metab. 33, 1655–1670. e8 (2021).
Li, Y. E. et al. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J. Cell Biol. 220, e202103105 (2021).
Matoba, K. et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 27, 1185–1193 (2020).
Ghanbarpour, A., Valverde, D. P., Melia, T. J. & Reinisch, K. M. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. USA 118, e2101562118 (2021).
Pastor-Cantizano, N., Montesinos, J. C., Bernat-Silvestre, C., Marcote, M. J. & Aniento, F. p24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma 253, 967–985 (2016).
Russell, R. C., Yuan, H. X. & Guan, K. L. Autophagy regulation by nutrient signaling. Cell Res. 24, 42–57 (2014).
Li, T. Y., Lin, S. Y. & Lin, S. C. Mechanism and physiological significance of growth factor-related autophagy. Physiology 28, 423–431 (2013).
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007).
Sun, N. et al. Measuring in vivo mitophagy. Mol. Cell 60, 685–696 (2015).
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Julien, J. P. & Kriz, J. Transgenic mouse models of amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1762, 1013–1024 (2006).
Ge, L. et al. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 18, 1586–1603 (2017).
Weisong, Z. et al. Research Square https://doi.org/10.21203/rs.3.rs-279271/v1 (2021).
Dominguez, M. et al. gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 140, 751–765 (1998).
Alford, S. C., Ding, Y., Simmen, T. & Campbell, R. E. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth. Biol. 1, 569–575 (2012).
Boncompain, G. et al. Synchronization of secretory protein traffic in populations of cells. Nat. Methods 9, 493–498 (2012).
Zanetti, G., Pahuja, K. B., Studer, S., Shim, S. & Schekman, R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14, 20–28 (2012).
Sawa-Makarska, J. et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 369, eaaz7714 (2020).
Zhang, M. et al. A translocation pathway for vesicle-mediated unconventional protein secretion. Cell 181, 637–652.e615 (2020).
Carinci, M. et al. TFG binds LC3C to regulate ULK1 localization and autophagosome formation. EMBO J. 40, e103563 (2021).
Graef, M., Friedman, J. R., Graham, C., Babu, M. & Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 24, 2918–2931 (2013).
Suzuki, K., Akioka, M., Kondo-Kakuta, C., Yamamoto, H. & Ohsumi, Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126, 2534–2544 (2013).
Tan, D. et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA 110, 19432–19437 (2013).
Lynch-Day, M. A. et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107, 7811–7816 (2010).
Kakuta, S. et al. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J. Biol. Chem. 287, 44261–44269 (2012).
Wang, J. et al. Ypt1/Rab1 regulates Hrr25/CK1delta kinase activity in ER-Golgi traffic and macroautophagy. J. Cell Biol. 210, 273–285 (2015).
Ding, X. et al. RAB2 regulates the formation of autophagosome and autolysosome in mammalian cells. Autophagy 15, 1774–1786 (2019).
Zoppino, F. C., Militello, R. D., Slavin, I., Alvarez, C. & Colombo, M. I. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11, 1246–1261 (2010).
Lamb, C. A. et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 35, 281–301 (2016).
Kurokawa, K., Okamoto, M. & Nakano, A. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 5, 3653 (2014).
Weigel, A. V. et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184, 2412–2429.e2416 (2021).
Shomron, O. et al. COPII collar defines the boundary between ER and ER exit site and does not coat cargo containers. J. Cell Biol. 220, e201907224 (2021).
Zeuschner, D. et al. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat. Cell Biol. 8, 377–383 (2006).
Venditti, R., Masone, M. C. & De Matteis, M. A. ER-Golgi membrane contact sites. Biochem. Soc. Trans. 48, 187–197 (2020).
Venditti, R. et al. Molecular determinants of ER-Golgi contacts identified through a new FRET-FLIM system. J. Cell Biol. 218, 1055–1065 (2019).
Venditti, R. et al. The activity of Sac1 across ER-TGN contact sites requires the four-phosphate-adaptor-protein-1. J. Cell Biol. 218, 783–797 (2019).
Schweizer, A. et al. Identification of an intermediate compartment involved in protein transport from endoplasmic reticulum to Golgi apparatus. Eur. J. Cell Biol. 53, 185–196 (1990).
Saraste, J. & Prydz, K. Assembly and cellular exit of coronaviruses: hijacking an unconventional secretory pathway from the pre-Golgi intermediate compartment via the Golgi ribbon to the extracellular space. Cells 10, 503 (2021).
Yan, R., Chen, K., Xu, K. Tubular ERGIC (t-ERGIC): a SURF4-mediated expressway for ER-to-Golgi transport. bioRxiv https://doi.org/10.1101/2021.04.06.438517 (2021).
Itakura, E., Kishi, C., Inoue, K. & Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 (2008).
Shoji-Kawata, S. et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013).
Yuan, L., Kenny, S. J., Hemmati, J., Xu, K. & Schekman, R. TANGO1 and SEC12 are copackaged with procollagen I to facilitate the generation of large COPII carriers. Proc. Natl. Acad. Sci. USA 115, E12255–E12264 (2018).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Appenzeller-Herzog, C. & Hauri, H. P. The ER-Golgi intermediate compartment (ERGIC): in search of its identity and function. J. Cell Sci. 119, 2173–2183 (2006).
Hughes, H. et al. Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J. Cell Sci. 122, 2924–2934 (2009).
Acknowledgements
L. G. is deeply grateful to Dr Randy Schekman for the post-doctoral training at UC Berkeley. We thank Dr Hongguang Xia (Zhejiang University, China), Dr Houchaima Ben-Tekaya (University of Basel, Switzerland), and Dr Yueguang Rong (Huazhong University of Science and Technology, China) for reagents, Peng Zhao and Huijiao Bai at Tsinghua University for technical assistance. We thank Dr Li Yu, Dr Ye-Guang Chen (Tsinghua University, China), Dr Wei Liu (Zhejiang University, China), Dr Quan Chen (Nankai University, China), Dr Hong Zhang (Institute of Biophysics, Chinese Academy of Sciences, China) and Dr Qing Zhong (Shanghai Jiao Tong University, China) for helpful suggestions on the study. The authors would like to acknowledge the assistance of Imaging Core Facility, Technology Center for Protein Sciences, Tsinghua University. The work is funded by National Key R&D Program of China (2019YFA0508602), the National Natural Science Foundation of China (91854114, 32061143009, 31872832, 31872826), Beijing Natural Science Foundation (JQ20028), Tsinghua Independent Research Program (2019Z06QCX02), Tsinghua University COVID-19 Emergency Aid Fund (2020Z99CFZ024), and Tsinghua-Peking Center for Life Sciences.
Author information
Authors and Affiliations
Contributions
Shulin L., and L.G. performed experiments, analyzed data, and wrote the manuscript. R.Y., J.X., S.Z., X.M., Y.L., and J.-J.G.L. performed experiments. M.Z., Q.S., L.C., Sai L., and K.X. analyzed data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, S., Yan, R., Xu, J. et al. A new type of ERGIC–ERES membrane contact mediated by TMED9 and SEC12 is required for autophagosome biogenesis. Cell Res 32, 119–138 (2022). https://doi.org/10.1038/s41422-021-00563-0
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41422-021-00563-0
This article is cited by
-
Towards a unified framework for the function of endoplasmic reticulum exit sites
Nature Reviews Molecular Cell Biology (2025)
-
A dual role of ERGIC-localized Rabs in TMED10-mediated unconventional protein secretion
Nature Cell Biology (2024)
-
C9orf72 controls hepatic lipid metabolism by regulating SREBP1 transport
Cell Death & Differentiation (2024)
-
Localized, highly efficient secretion of signaling proteins by migrasomes
Cell Research (2024)
-
Biomolecular condensates mediate bending and scission of endosome membranes
Nature (2024)