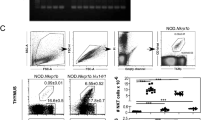
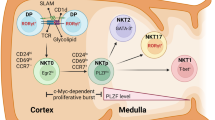
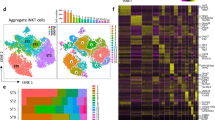
Abstract
Invariant natural killer T (iNKT) cells develop from CD4+CD8+ double-positive (DP) thymocytes and express an invariant Vα14–Jα18 T-cell receptor (TCR) α-chain. Generation of these cells requires the prolonged survival of DP thymocytes to allow for Vα14–Jα18 gene rearrangements and strong TCR signaling to induce the expression of the iNKT lineage-specific transcription factor PLZF. Here, we report that the transcription factor Yin Yang 1 (YY1) is essential for iNKT cell formation. Thymocytes lacking YY1 displayed a block in iNKT cell development at the earliest progenitor stage. YY1-deficient thymocytes underwent normal Vα14–Jα18 gene rearrangements, but exhibited impaired cell survival. Deletion of the apoptotic protein BIM failed to rescue the defect in iNKT cell generation. Chromatin immunoprecipitation and deep-sequencing experiments demonstrated that YY1 directly binds and activates the promoter of the Plzf gene. Thus, YY1 plays essential roles in iNKT cell development by coordinately regulating cell survival and PLZF expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Makino, Y., Kanno, R., Ito, T., Higashino, K. & Taniguchi, M. Predominant expression of invariant V alpha 14+ TCR alpha chain in NK1.1+ T cell populations. Int. Immunol. 7, 1157–1161 (1995).
Fowlkes, B. J. et al. A novel population of T-cell receptor alpha beta-bearing thymocytes which predominantly expresses a single V beta gene family. Nature 329, 251–254 (1987).
Ceredig, R., Lynch, F. & Newman, P. Phenotypic properties, interleukin 2 production, and developmental origin of a “mature” subpopulation of Lyt-2- L3T4- mouse thymocytes. Proc. Natl Acad. Sci. USA 84, 8578–8582 (1987).
Budd, R. C. et al. Developmentally regulated expression of T cell receptor beta chain variable domains in immature thymocytes. J. Exp. Med. 166, 577–582 (1987).
Salio, M., Silk, J. D., Jones, E. Y. & Cerundolo, V. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol. 32, 323–366 (2014).
Godfrey, D. I. & Berzins, S. P. Control points in NKT-cell development. Nat. Rev. Immunol. 7, 505–518 (2007).
Guo, J. et al. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 3, 469–476 (2002).
Hu, T., Simmons, A., Yuan, J., Bender, T. P. & Alberola-Ila, J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat. Immunol. 11, 435–441 (2010).
Callen, E. et al. The DNA damage- and transcription-associated protein paxip1 controls thymocyte development and emigration. Immunity 37, 971–985 (2012).
D’Cruz, L. M., Knell, J., Fujimoto, J. K. & Goldrath, A. W. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat. Immunol. 11, 240–249 (2010).
Egawa, T. et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008).
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Lazarevic, V. et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10, 306–313 (2009).
Seiler, M. P. et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13, 264–271 (2012).
Dose, M. et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl Acad. Sci. USA 106, 8641–8646 (2009).
Mycko, M. P. et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J. Immunol. 182, 4641–4648 (2009).
Walunas, T. L., Wang, B., Wang, C. R. & Leiden, J. M. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J. Immunol. 164, 2857–2860 (2000).
Townsend, M. J. et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20, 477–494 (2004).
Monticelli, L. A. et al. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc. Natl Acad. Sci. USA 106, 19461–19466 (2009).
van Gisbergen, K. P. et al. Mouse Hobit is a homolog of the transcriptional repressor Blimp-1 that regulates NKT cell effector differentiation. Nat. Immunol. 13, 864–871 (2012).
Kim, P. J. et al. GATA-3 regulates the development and function of invariant NKT cells. J. Immunol. 177, 6650–6659 (2006).
Lacorazza, H. D. et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17, 437–449 (2002).
Stanic, A. K. et al. Cutting edge: the ontogeny and function of Va14Ja18 natural T lymphocytes require signal processing by protein kinase C theta and NF-kappa B. J. Immunol. 172, 4667–4671 (2004).
Flanagan, J. R. et al. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol. Cell. Biol. 12, 38–44 (1992).
Shi, Y., Seto, E., Chang, L. S. & Shenk, T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67, 377–388 (1991).
Park, K. & Atchison, M. L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3’ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc. Natl Acad. Sci. USA 88, 9804–9808 (1991).
Hariharan, N., Kelley, D. E. & Perry, R. P. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc. Natl Acad. Sci. USA 88, 9799–9803 (1991).
Kleiman, E., Jia, H., Loguercio, S., Su, A. I. & Feeney, A. J. YY1 plays an essential role at all stages of B-cell differentiation. Proc. Natl Acad. Sci. USA 113, E3911–E3920 (2016).
Liu, H. et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 21, 1179–1189 (2007).
Trabucco, S. E., Gerstein, R. M. & Zhang, H. YY1 regulates the germinal center reaction by inhibiting apoptosis. J. Immunol. 197, 1699–1707 (2016).
Banerjee, A. et al. YY1 is required for germinal center B cell development. PLoS ONE 11, e0155311 (2016).
Green, M. R. et al. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl Acad. Sci. USA 108, 2873–2878 (2011).
Hwang, S. S. et al. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc. Natl Acad. Sci. USA 110, 276–281 (2013).
Yu, B. et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat. Immunol. 18, 573–582 (2017).
Hwang, S. S. et al. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat. Commun. 7, 10789 (2016).
Kwon, H. K., Chen, H. M., Mathis, D. & Benoist, C. Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat. Immunol. 18, 1238–1248 (2017).
Wang, G. Z. & Goff, S. P. Regulation of Yin Yang 1 by tyrosine phosphorylation. J. Biol. Chem. 290, 21890–21900 (2015).
Ou, X., Xu, S., Li, Y. F. & Lam, K. P. Adaptor protein DOK3 promotes plasma cell differentiation by regulating the expression of programmed cell death 1 ligands. Proc. Natl Acad. Sci. USA 111, 11431–11436 (2014).
Potluri, V. et al. Transcriptional repression of Bim by a novel YY1-RelA complex is essential for the survival and growth of multiple myeloma. PLoS ONE 8, e66121 (2013).
He, Y. et al. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat. Neurosci. 13, 1472–1480 (2010).
Lu, L. et al. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 32, 2575–2588 (2013).
Chen, L. et al. Genome-wide analysis of YY2 versus YY1 target genes. Nucleic Acids Res. 38, 4011–4026 (2010).
Chen, L., Foreman, D. P., Sant’Angelo, D. B. & Krangel, M. S. Yin Yang 1 promotes thymocyte survival by downregulating p53. J. Immunol. 196, 2572–2582 (2016).
Bezbradica, J. S., Hill, T., Stanic, A. K., Van Kaer, L. & Joyce, S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc. Natl Acad. Sci. USA 102, 5114–5119 (2005).
Gapin, L., Matsuda, J. L., Surh, C. D. & Kronenberg, M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2, 971–978 (2001).
Acknowledgements
We thank Ms Lim Wei Lee for sample processing and plasmid construction. We also thank other members of the laboratory for insightful discussion and the A*STAR Biomedical Research Council for grant support.
Author information
Authors and Affiliations
Contributions
X.O., S.X., and K.-P.L. designed the experiments and wrote the paper. X.O., J.H., Y.H., and Y.-F.L. conducted the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ou, X., Huo, J., Huang, Y. et al. Transcription factor YY1 is essential for iNKT cell development. Cell Mol Immunol 16, 547–556 (2019). https://doi.org/10.1038/s41423-018-0002-6
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41423-018-0002-6
Keywords
This article is cited by
-
YY1 regulates the proliferation and invasion of triple-negative breast cancer via activating PLAUR
Functional & Integrative Genomics (2023)