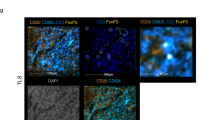
Abstract
Regulatory T cells (Tregs) establish dominant immune tolerance but obstruct tumor immune surveillance, warranting context-specific mechanistic insights into the functions of tumor-infiltrating Tregs (TIL-Tregs). We show that enhanced posttranslational O-linked N-acetylglucosamine modification (O-GlcNAcylation) of cellular factors is a molecular feature that promotes a tumor-specific gene expression signature and distinguishes TIL-Tregs from their systemic counterparts. We found that altered glucose utilization through the glucose transporter Glut3 is a major facilitator of this process. Treg-specific deletion of Glut3 abrogates tumor immune tolerance, while steady-state immune homeostasis remains largely unaffected in mice. Furthermore, by employing mouse tumor models and human clinical data, we identified the NF-κB subunit c-Rel as one such factor that, through Glut3-dependent O-GlcNAcylation, functionally orchestrates gene expression in Tregs at tumor sites. Together, these results not only identify immunometabolic alterations and molecular events contributing to fundamental aspects of Treg biology, specifically at tumor sites but also reveal tumor-specific cellular properties that can aid in the development of Treg-targeted cancer immunotherapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The RNA-seq datasets that support the findings of this study are deposited in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE235402.
References
Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–64.
Tanaka A, Sakaguchi S. Targeting Treg cells in cancer immunotherapy. Eur J Immunol. 2019;49:1140–6.
Liu C, Chikina M, Deshpande R, Menk AV, Wang T, Tabib T, et al. Treg Cells Promote the SREBP1-Dependent Metabolic Fitness of Tumor-Promoting Macrophages via Repression of CD8(+) T-Cell-Derived Interferon-gamma. Immunity. 2019;51:381–97.e6.
Bos P, Rudensky A. Transient regulatory T-cell ablation deters oncogene driven breast cancer and enhances radiotherapy. J Exp Med. 2013;210:2435–46.
Klages K, Mayer CT, Lahl K, Loddenkemper C, Teng MW, Ngiow SF, et al. Selective depletion of Foxp3+ regulatory T cells improves effective therapeutic vaccination against established melanoma. Cancer Res. 2010;70:7788–99.
Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, Ohkura N. Regulatory T Cells and Human Disease. Annu Rev Immunol. 2020;38:541–66.
Watson MJ, Vignali PDA, Mullett SJ, Overacre-Delgoffe AE, Peralta RM, Grebinoski S, et al. Metabolic support of tumor-infiltrating regulatory T cells by lactic acid. Nature. 2021;591:645–51.
Zappasodi R, Serganova I, Cohen IJ, Maeda M, Shindo M, Senbabaoglu Y, et al. CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumors. Nature. 2021;591:652–8.
Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 Signaling Pathway Regulates Glucose Metabolism. Immunity. 2002;16:769–77.
Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T-cell activation and effector function. Cell Metab. 2014;20:61–72.
Ho PC, Kaech SM. Reenergizing T-cell anti-tumor immunity by harnessing immunometabolic checkpoints and machineries. Curr Opin Immunol. 2017;46:38–44.
Simpson IA, Dwyer D, Malide D, Moley KH, Travis A, Vannucci SJ. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Endocrinol Metab. 2008;295:E242–53.
Hochrein SM, Wu H, Eckstein M, Arrigoni L, Herman JS, Schumacher F, et al. The glucose transporter GLUT3 controls T helper 17 cell responses through glycolytic-epigenetic reprogramming. Cell Metab. 2022;34:516–32.e11.
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W14.
Magnuson AM, Kiner E, Ergun A, Park JS, Asinovski N, Ortiz-Lopez A, et al. Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci USA. 2018;115:E10672–E81.
Sercan O, Stoycheva D, Hammerling GJ, Arnold B, Schuler T. IFN-gamma receptor signaling regulates memory CD8+ T-cell differentiation. J Immunol. 2010;184:2855–62.
Zheng L, Fisher G, Miller RE, Peschon J, Lynch DH, Lenardo MJ. Induction of apoptosis in mature T cells by tumor necrosis factor. Nature. 1995;377:348–51.
Bollyky PL, Falk BA, Long SA, Preisinger A, Braun KR, Wu RP, et al. CD44 costimulation promotes FoxP3+ regulatory T-cell persistence and function via production of IL-2, IL-10, and TGF-beta. J Immunol. 2009;183:2232–41.
Flynn KM, Michaud M, Madri JA. CD44 deficiency contributes to enhanced experimental autoimmune encephalomyelitis: a role in immune cells and vascular cells of the blood‒brain barrier. Am J Pathol. 2013;182:1322–36.
de Oliveira CE, Gasparoto TH, Pinheiro CR, Amor NG, Nogueira MRS, Kaneno R, et al. CCR5-Dependent Homing of T Regulatory Cells to the Tumor Microenvironment Contributes to Skin Squamous Cell Carcinoma Development. Mol Cancer Ther. 2017;16:2871–80.
Xia S, Liu X, Cao X, Xu S. T-cell expression of Bruton’s tyrosine kinase promotes autoreactive T-cell activation and exacerbates aplastic anemia. Cell Mol Immunol. 2020;17:1042–52.
Trzupek D, Dunstan M, Cutler AJ, Lee M, Godfrey L, Jarvis L, et al. Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein‒RNA single-cell analysis. Genome Med. 2020;12:55.
Sutmuller RP, den Brok MH, Kramer M, Bennink EJ, Toonen LW, Kullberg BJ, et al. Toll-like receptor 2 controls expansion and function of regulatory T cells. J Clin Invest. 2006;116:485–94.
Grinberg-Bleyer Y, Oh H, Desrichard A, Bhatt DM, Caron R, Chan TA, et al. NF-kappaB c-Rel Is Crucial for the Regulatory T-Cell Immune Checkpoint in Cancer. Cell. 2017;170:1096–108.e13.
Oh H, Grinberg-Bleyer Y, Liao W, Maloney D, Wang P, Wu Z, et al. An NF-kappaB Transcription-Factor-Dependent Lineage-Specific Transcriptional Program Promotes Regulatory T-Cell Identity and Function. Immunity. 2017;47:450–65.e5.
Arvey A, van der Veeken J, Samstein RM, Feng Y, Stamatoyannopoulos JA, Rudensky AY. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2014;15:580–7.
Lee JB, Pyo KH, Kim HR. Role and Function of O-GlcNAcylation in Cancer. Cancers (Basel). 2021;13:5365–79.
Shimizu M, Tanaka N. IL-8-induced O-GlcNAc modification via GLUT3 and GFAT regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene. 2019;38:1520–33.
Gu J, Jin N, Ma D, Chu D, Iqbal K, Gong CX, et al. Calpain I Activation Causes GLUT3 Proteolysis and Downregulation of O-GlcNAcylation in Alzheimer’s Disease Brain. J Alzheimers Dis. 2018;62:1737–46.
Ramakrishnan P, Clark PM, Mason DE, Peters EC, Hsieh-Wilson LC, Baltimore D. Activation of the transcriptional function of the NF-kappaB protein c-Rel by O-GlcNAc glycosylation. Sci Signal. 2013;6:ra75.
Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–9.
Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved noncoding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–12.
Lanczky A, Gyorffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med internet Res. 2021;23:e27633.
Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, et al. CD36-mediated metabolic adaptation supports regulatory T-cell survival and function in tumors. Nat Immunol. 2020;21:298–308.
Bottcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022–37.e14.
Zhang Z, Tan EP, VandenHull NJ, Peterson KR, Slawson C. O-GlcNAcase Expression is Sensitive to Changes in O-GlcNAc Homeostasis. Front Endocrinol (Lausanne). 2014;5:206.
Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011;7:174–81.
Kazemi Z, Chang H, Haserodt S, McKen C, Zachara NE. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3beta-dependent manner. J Biol Chem. 2010;285:39096–107.
Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18:452–65.
Liu B, Salgado OC, Singh S, Hippen KL, Maynard JC, Burlingame AL, et al. The lineage stability and suppressive program of regulatory T cells require protein O-GlcNAcylation. Nat Commun. 2019;10:354.
Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, et al. Metabolic programming and PDHK1 control CD4+ T-cell subsets and inflammation. J Clin Invest. 2015;125:194–207.
Reinfeld BI, Madden MZ, Wolf MM, Chytil A, Bader JE, Patterson AR, et al. Cell-programmed nutrient partitioning in the tumor microenvironment. Nature. 2021;593:282–8.
Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T-Cell Responses. Cell. 2015;162:1217–28.
Priyadharshini B, Loschi M, Newton RH, Zhang JW, Finn KK, Gerriets VA, et al. Cutting Edge: TGF-beta and Phosphatidylinositol 3-Kinase Signals Modulate Distinct Metabolism of Regulatory T-Cell Subsets. J Immunol. 2018.
De Rosa V, Galgani M, Porcellini A, Colamatteo A, Santopaolo M, Zuchegna C, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat Immunol. 2015;16:1174–84.
Procaccini C, Carbone F, Di Silvestre D, Brambilla F, De Rosa V, Galgani M, et al. The Proteomic Landscape of Human Ex Vivo Regulatory and Conventional T Cells Reveals Specific Metabolic Requirements. Immunity. 2016;44:406–21.
Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, et al. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat Immunol. 2016;17:1459–66.
Tanner LB, Goglia AG, Wei MH, Sehgal T, Parsons LR, Park JO, et al. Four Key Steps Control Glycolytic Flux in Mammalian Cells. Cell Syst. 2018;7:49–62.e8.
Cosset E, Ilmjarv S, Dutoit V, Elliott K, von Schalscha T, Camargo MF, et al. Glut3 Addiction Is a Druggable Vulnerability for a Molecularly Defined Subpopulation of Glioblastoma. Cancer Cell. 2017;32:856–68.e5.
Hwang SY, Hwang JS, Kim SY, Han IO. O-GlcNAcylation and p50/p105 binding of c-Rel are dynamically regulated by LPS and glucosamine in BV2 microglia cells. Br J Pharm. 2013;169:1551–60.
Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J. 2007;26:4368–79.
Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DM, et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T-cell self-renewal and malignancy. Nat Immunol. 2016;17:712–20.
Saravia J, Zeng H, Dhungana Y, Bastardo Blanco D, Nguyen TM, Chapman NM, et al. Homeostasis and transitional activation of regulatory T cells require c-Myc. Sci Adv. 2020;6:eaaw6443.
Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800.
Cribioli E, Giordano Attianese GMP, Ginefra P, Signorino-Gelo A, Vuillefroy de Silly R, Vannini N, et al. Enforcing GLUT3 expression in CD8(+) T cells improves fitness and tumor control by promoting glucose uptake and energy storage. Front Immunol. 2022;13:976628.
Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–7.
Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, et al. Regulatory T-cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–58.
Liston A, Nutsch KM, Farr AG, Lund JM, Rasmussen JP, Koni PA, et al. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc Natl Acad Sci USA. 2008;105:11903–8.
Loo CS, Gatchalian J, Liang Y, Leblanc M, Xie M, Ho J, et al. A Genome-wide CRISPR Screen Reveals a Role for the Noncanonical Nucleosome-Remodeling BAF Complex in Foxp3 Expression and Regulatory T-Cell Function. Immunity. 2020;53:143–57.e8.
Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–5.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Fu G, Guy CS, Chapman NM, Palacios G, Wei J, Zhou P, et al. Metabolic control of TFH cells and humoral immunity by phosphatidylethanolamine. Nature. 2021;595:724–9.
Kamburov A, Cavill R, Ebbels TM, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–8.
Hegazy M, Cohen-Barak E, Koetsier JL, Najor NA, Arvanitis C, Sprecher E, et al. Proximity Ligation Assay for Detecting Protein‒Protein Interactions and Protein Modifications in Cells and Tissues in Situ. Curr Protoc Cell Biol. 2020;89:e115.
Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556–W60.
Chowdhury S, Kar A, Bhowmik D, Gautam A, Basak D, Sarkar I, et al. Intracellular Acetyl-CoA Potentiates the Therapeutic Efficacy of Antitumor CD8+ T Cells. Cancer Res. 2022;82:2640–55.
Shen L, Hu P, Zhang Y, Ji Z, Shan X, Ni L, et al. Serine metabolism antagonizes antiviral innate immunity by preventing ATP6V0d2-mediated YAP lysosomal degradation. Cell Metab. 2021;33:971–87.e6.
Zimmermann M, Sauer U, Zamboni N. Quantification and mass isotopomer profiling of alpha-keto acids in central carbon metabolism. Anal Chem. 2014;86:3232–7.
Tian H, Ni Z, Lam SM, Jiang W, Li F, Du J, et al. Precise Metabolomics Reveals a Diversity of Aging-Associated Metabolic Features. Small Methods. 2022;6:e2200130.
Acknowledgements
We thank H. Jung and H. Nam for their technical assistance in cell sorting and K. Cho for their technical assistance in confocal microscopy. We also acknowledge the assistance of Dr. Gustavo Palacios for lipidomic and metabolomic analysis. YC was financially supported by the Dissertation Completion Award of the University of Georgia and American Lebanese Syrian Associated Charities (ALSAC) at St. Jude Children’s Research Hospital. AG acknowledges support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through grant no INST 37/935-1 FUGG, and the de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) and ELIXIR-DE (Forschungszentrum Jülich and W-de.NBI-001, W-de.NBI-004, W-de.NBI-008, W-de.NBI-010, W-de.NBI-013, W-de.NBI-014, W-de.NBI-016, W-de.NBI-022) for supporting the computational analysis carried out in this work. AS was supported by BrainKorea21 Plus scholarship from the National Research Foundation of Korea (NRF). This research, in part, was supported by the Basic Science Research Program (grant# 4.24643.01) funded by the Ministry of Education, Korea, and NRF grants# RS-2023–00260454 (AS) and RS-2024–00345575 (SHI) funded by the Korea Ministry of Science and ICT (MSIT). DR is recipient of National Natural Science Foundation of China grant# 32470980.
Author information
Authors and Affiliations
Contributions
AS, SHI, and DR conceptualized the project and devised the project outline. AS performed most of the experiments, analyzed the data, and wrote the manuscript. Garima Sharma performed the animal tumor experiments, flow cytometry and cell sorting. ZG performed the EAE and cell transfer-induced colitis experiments. KL and ML performed the animal tumor experiments and in vitro iTreg-based metabolic flux and 13C6-labeling experiments. MW standardized and prepared the tumor-conditioned media for metabolic flux experiments. CJK processed the human tissue sections for PLA. YC performed and analyzed the metabolomic data. AG analyzed and visualized the metabolomics data. HBC, JK, and JMK provided human tumor tissue samples. SML and Guanghou Shui performed sample preparation and LC‒MS for 13C6-labeling experiments. SP directed the analysis of metabolomic data. YF provided scientific insights and facilities for exploratory metabolomic analysis. KK performed the computational analysis of the RNA-Seq data. SHI and DR directed the project, obtained funding, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Garima Sharma is an employee of ImmunoBiome Inc, South Korea. SHI is the founder and major shareholder of ImmunoBiome Inc, South Korea. SML is an employee of Lipidall Technologies Company Limited, China. The authors have no conflicting financial interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sharma, A., Sharma, G., Gao, Z. et al. Glut3 promotes cellular O-GlcNAcylation as a distinctive tumor-supportive feature in Treg cells. Cell Mol Immunol 21, 1474–1490 (2024). https://doi.org/10.1038/s41423-024-01229-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41423-024-01229-8
Keywords
This article is cited by
-
GLUT1 expression, lymphocyte distribution and CD3+ T-cell metabolic subsets as predictive markers of response to immunotherapy in advanced melanoma
Journal of Experimental & Clinical Cancer Research (2026)
-
Metabolic checkpoints in immune cell reprogramming: rewiring immunometabolism for cancer therapy
Molecular Cancer (2025)
-
Immunometabolism: crosstalk with tumor metabolism and implications for cancer immunotherapy
Molecular Cancer (2025)
-
A special RELationship between sugar and tumor-infiltrating regulatory T cells
Cellular & Molecular Immunology (2024)