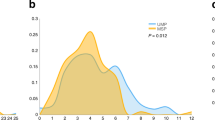
Abstract
Human leukocyte antigen (HLA) disparity between donors and recipients is a key determinant triggering intense alloreactivity, leading to a lethal complication, namely, acute graft-versus-host disease (aGVHD), after allogeneic transplantation. Moreover, aGVHD remains a cause of mortality after HLA-matched allogeneic transplantation. Protocols for HLA-haploidentical hematopoietic cell transplantation (haploHCT) have been established successfully and widely applied, further highlighting the urgency of performing panoramic screening of non-HLA variations correlated with aGVHD. On the basis of our time-consecutive large haploHCT cohort (with a homogenous discovery set and an extended confirmatory set), we first delineated the genetic landscape of 1366 samples to quantitatively model aGVHD risk by assessing the contributions of HLA and non-HLA genes together with clinical factors. In addition to identifying multiple loss-of-function (LoF) risk variations in non-HLA coding genes, our data-driven study revealed that non-HLA genetic variations, independent of HLA disparity, contributed the most to the occurrence of aGVHD. This unexpected major effect was verified in an independent cohort that received HLA-identical sibling HCT. Subsequent functional experiments further revealed the roles of a representative non-HLA LoF gene and LoF gene pair in regulating the alloreactivity of primary human T cells. Our findings highlight the importance of non-HLA genetic risk in the new era of transplantation and propose a new direction to explore the immunogenetic mechanism of alloreactivity and to optimize donor selection strategies for allogeneic transplantation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The raw data have been deposited at GSA (https://ngdc.cncb.ac.cn/gsa/) under the accession number PRJCA009965. All custom scripts used for analyses are available online (http://agvhd.gao-lab.org/). Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Xiao-Jun Huang.
References
Álvaro-Benito M, Morrison E, Ebner F, Abualrous ET, Urbicht M, Wieczorek M, et al. Distinct editing functions of natural HLA-DM allotypes impact antigen presentation and CD4+ T cell activation. Cell Mol Immunol. 2020;17:133–42.
Zeiser R, Teshima T. Nonclassical manifestations of acute GVHD. Blood. 2021;138:2165–72.
Velardi A, Mancusi A, Ruggeri L, Pierini A. How adoptive transfer of components of the donor immune system boosts GvL and prevents GvHD in HLA-haploidentical hematopoietic transplantation for acute leukemia. Bone Marrow Transpl. 2024;59:301–5.
Raiola AM, Risitano A, Sacchi N, Giannoni L, Signori A, Aquino S, et al. Impact of HLA disparity in haploidentical bone marrow transplantation followed by high-dose cyclophosphamide. Biol Blood Marrow Transpl. 2018;24:119–26.
de Almeida GP, Lichtner P, Eckstein G, Brinkschmidt T, Chu C-F, Sun S, et al. Human skin-resident host T cells can persist long term after allogeneic stem cell transplantation and maintain recirculation potential. Sci Immunol. 2022;7:eabe2634.
Galli E, Metafuni E, Giammarco S, Limongiello MA, Innocenti I, Autore F, et al. Triple post transplant cyclophosphamide (PTCY) based GVHD prophylaxis: HLA matched versus HLA haploidentical transplants. Bone Marrow Transpl. 2022;57:532–7.
Wang Y, Chang Y-J, Xu L-P, Liu K-Y, Liu D-H, Zhang X-H, et al. Who is the best donor for a related HLA haplotype-mismatched transplant? Blood. 2014;124:843–50.
Chang Y-J, Xu L-P, Wang Y, Zhang X-H, Chen H, Chen Y-H, et al. Controlled, randomized, open-label trial of risk-stratified corticosteroid prevention of acute graft-versus-host disease after haploidentical transplantation. J Clin Oncol. 2016;34:1855–63.
Kanakry CG, Fuchs EJ, Luznik L. Modern approaches to HLA-haploidentical blood or marrow transplantation. Nat Rev Clin Oncol. 2016;13:10–24.
Robinson TM, O’Donnell PV, Fuchs EJ, Luznik L. Haploidentical bone marrow and stem cell transplantation: experience with posttransplantation cyclophosphamide. Semin Hematol. 2016;53:90–7.
Yu S, Huang F, Fan Z, Xuan L, Nie D, Xu Y, et al. Haploidentical versus HLA-matched sibling transplantation for refractory acute leukemia undergoing sequential intensified conditioning followed by DLI: an analysis from two prospective data. J Hematol Oncol. 2020;13:18.
Robin M, Porcher R, Ruggeri A, Blaise D, Wolschke C, Koster L, et al. HLA-Mismatched Donors in Patients with Myelodysplastic Syndrome: An EBMT Registry Analysis. Biol Blood Marrow Transpl. 2019;25:114–20.
Lv M, Guo H-D, Huang X-J. A perfect mismatch: haploidentical hematopoietic stem cell transplantation overtakes a bend. Cell Mol Immunol. 2023;20:978–80.
Huo M-R, Pei X-Y, Li D, Chang Y-J, Xu L-P, Zhang X-H, et al. Impact of HLA allele mismatch at HLA-A, -B, -C, -DRB1, and -DQB1 on outcomes in haploidentical stem cell transplantation. Bone Marrow Transpl. 2018;53:600–8.
Kasamon YL, Luznik L, Leffell MS, Kowalski J, Tsai H-L, Bolaños-Meade J, et al. Nonmyeloablative HLA-haploidentical bone marrow transplantation with high-dose posttransplantation cyclophosphamide: effect of HLA disparity on outcome. Biol Blood Marrow Transpl. 2010;16:482–9.
Dickinson AM, Norden J. Non-HLA genomics: does it have a role in predicting hematopoietic stem cell transplantation outcome? Int J Immunogenet. 2015;42:229–38.
Goussetis E, Varela I, Peristeri I, Kitra V, Spanou K, Moraloglou O, et al. Cytokine gene polymorphisms and graft-versus-host disease in children after matched sibling hematopoietic stem cell transplantation: a single-center experience. Cell Mol Immunol. 2011;8:276–80.
Cieri N, Hookeri N, Stromhaug K, Li L, Keating J, Díaz-Fernández P, et al. Systematic identification of minor histocompatibility antigens predicts outcomes of allogeneic hematopoietic cell transplantation. Nat Biotechnol. 2024. https://doi.org/10.1038/s41587-024-02348-3. Online ahead of print.
Martin PJ, Levine DM, Storer BE, Warren EH, Zheng X, Nelson SC, et al. Genome-wide minor histocompatibility matching as related to the risk of graft-versus-host disease. Blood. 2017;129:791–8.
Cantoni N, Hirsch HH, Khanna N, Gerull S, Buser A, Bucher C, et al. Evidence for a bidirectional relationship between cytomegalovirus replication and acute graft-versus-host disease. Biol Blood Marrow Transpl. 2010;16:1309–14.
Akahoshi Y, Kimura S-I, Tada Y, Matsukawa T, Tamaki M, Doki N, et al. Cytomegalovirus gastroenteritis in patients with acute graft-versus-host disease. Blood Adv. 2022;6:574–84.
Riesner K, Cordes S, Peczynski C, Kalupa M, Schwarz C, Shi Y, et al. Reduced calcium signaling is associated with severe graft-versus-host disease: results from preclinical models and from a prospective EBMT Study. Front Immunol. 2020;11:1983.
Ogasawara R, Hashimoto D, Kimura S, Hayase E, Ara T, Takahashi S, et al. Intestinal lymphatic endothelial cells produce R-Spondin3. Sci Rep. 2018;8:10719.
Hayase E, Hashimoto D, Nakamura K, Noizat C, Ogasawara R, Takahashi S, et al. R-Spondin1 expands Paneth cells and prevents dysbiosis induced by graft-versus-host disease. J Exp Med. 2017;214:3507–18.
Lu C, Ma H, Song L, Wang H, Wang L, Li S, et al. IFN-γR/STAT1 signaling in recipient hematopoietic antigen-presenting cells suppresses graft-versus-host disease. J Clin Invest. 2023;133:e125986.
Zhang Y, Shen L, Dreißigacker K, Zhu H, Trinh-Minh T, Meng X, et al. Targeting of canonical WNT signaling ameliorates experimental sclerodermatous chronic graft-versus-host disease. Blood. 2021;137:2403–16.
Rakotomamonjy A. Variable selection using SVM based criteria. J Mach Learn Res. 2003;3:1357–70.
Liu QZ, Chen CH, Zhang Y, Hu ZG. Feature selection for support vector machines with RBF kernel. Artif Intell Rev. 2011;36:99–115.
Zhao JQ, Karimzadeh M, Masjedi A, Wang TJ, Zhang XW, Crawford MM, et al. FeatureExplorer: Interactive Feature Selection and Exploration of Regression Models for Hyperspectral Images. 2019 IEEE Visualization Conference (Vis). 2019:161–5.
Ciurea SO. Considerations for haploidentical versus unrelated donor transplants. Bone Marrow Transpl. 2019;54:738–42.
Sanz J, Labopin M, Choi G, Kulagin A, Peccatori J, Vydra J, et al. Younger unrelated donors may be preferable over HLA match in the PTCy era: a study from the ALWP of the EBMT. Blood. 2024;143:2534–43.
Sato-Otsubo A, Nannya Y, Kashiwase K, Onizuka M, Azuma F, Akatsuka Y, et al. Genome-wide surveillance of mismatched alleles for graft-versus-host disease in stem cell transplantation. Blood. 2015;126:2752–63.
Harkensee C, Oka A, Onizuka M, Middleton PG, Inoko H, Hirayasu K, et al. Single nucleotide polymorphisms and outcome risk in unrelated mismatched hematopoietic stem cell transplantation: an exploration study. Blood. 2012;119:6365–72.
Wang W, Huang H, Halagan M, Vierra-Green C, Heuer M, Brelsford JE, et al. Chromosome Y-encoded antigens associate with acute graft-versus-host disease in sex-mismatched stem cell transplant. Blood Adv. 2018;2:2419–29.
Chien JW, Zhang XC, Fan W, Wang H, Zhao LP, Martin PJ, et al. Evaluation of published single nucleotide polymorphisms associated with acute GVHD. Blood. 2012;119:5311–9.
Paczesny S. Biomarkers for posttransplantation outcomes. Blood. 2018;131:2193–204.
Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–7.
Álvaro-Benito M. Natural variation of ncHLAII molecules: challenges and perspectives. Cell Mol Immunol. 2022;19:1432–4.
Ma Z, Mao C, Jia Y, Yu F, Xu P, Tan Y, et al. ADAMTS7-Mediated complement factor H degradation potentiates complement activation to contributing to renal injuries. J Am Soc Nephrol. 2023;34:291–308.
Torices S, Alvarez-Rodríguez L, Grande L, Varela I, Muñoz P, Pascual D, et al. A Truncated Variant of ASCC1, a Novel Inhibitor of NF-κB, Is associated with disease severity in patients with rheumatoid arthritis. J Immunol. 2015;195:5415–20.
Fontalba A, Martinez-Taboada V, Gutierrez O, Pipaon C, Benito N, Balsa A, et al. Deficiency of the NF-kappaB inhibitor caspase activating and recruitment domain 8 in patients with rheumatoid arthritis is associated with disease severity. J Immunol. 2007;179:4867–73.
Kim DH, Lee B, Lee J, Kim ME, Lee JS, Chung JH, et al. FoxO6-mediated IL-1β induces hepatic insulin resistance and age-related inflammation via the TF/PAR2 pathway in aging and diabetic mice. Redox Biol. 2019;24:101184.
Kabir AU, Subramanian M, Lee DH, Wang X, Krchma K, Wu J, et al. Dual role of endothelial Myct1 in tumor angiogenesis and tumor immunity. Sci Transl Med. 2021;13:eabb6731.
Cros J, Cagnard N, Woollard K, Patey N, Zhang S-Y, Senechal B, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–86.
Murphy WJ. Drilling down interferon in GVHD/GVL. Blood. 2023;141:821–3.
Zheng X, Tian Z. Which is better, HLA-matched sibling or haploidentical transplantation? Cell Mol Immunol. 2021;18:1347.
Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21:2409–19.
Puhr R, Heinze G, Nold M, Lusa L, Geroldinger A. Firth’s logistic regression with rare events: accurate effect estimates and predictions? Stat Med. 2017;36:2302–17.
Anderson MJ, Legendre P. An empirical comparison of permutation methods for tests of partial regression coefficients in a linear model. J Stat Comput Simul. 1999;62:271–303.
Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. Neuroimage. 2014;92:381–97.
Anderson MJ, Robinson J. Permutation tests for linear models. Aust NZ J Stat. 2001;43:75–88.
Acknowledgements
This work was supported by the Major Program of the National Natural Science Foundation of China (No. 82293630), the Peking University Medicine Fund for the world’s leading discipline or discipline cluster development (No. 71003Y3035), the National Key Research and Development Program of China (Nos. 2022YFA0103300, 2017YFA0104500, and 2016YFC0901603), and the State Key Laboratory of Gene Function and Modulation Research and the Beijing Advanced Innovation Center for Genomics (ICG) at Peking University.
Author information
Authors and Affiliations
Contributions
SL, Y-JK, JL, GG, and X-JH: study design and project coordination. SL, MH, ML, and KY: cohort and sample collection and experimental design. YW, L-PX, X-HZ, NW, RL, and XD: cohort and sample collection. Y-JK, SL, MH, D-CY, C-RX, J-YL, JL, GG, and X-JH: data analyses. D-CY and Y-JK: web server development. SL, Y-JK, and JL: writing–original draft. JL, GG, and X-JH: writing–review & editing. JL, GG, and X-JH: project administration.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, S., Kang, YJ., Huo, M. et al. Systematic mining and quantification reveal the dominant contribution of non-HLA variations to acute graft-versus-host disease. Cell Mol Immunol 22, 501–511 (2025). https://doi.org/10.1038/s41423-025-01273-y
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41423-025-01273-y