Abstract
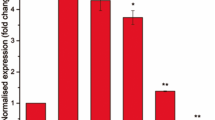
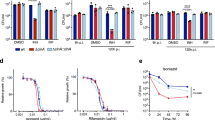
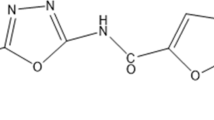
Isoniazid (INH) is one among the four first-line drugs used in the treatment of tuberculosis. The bactericidal activity of INH is due to its ability to inhibit mycolic acid synthesis, which is an integral component of the mycobacterial cell wall. Non-replicating Mycobacterium tuberculosis (MTB) is phenotypically resistant to INH. The exact mechanism of this resistance is not clear, although the inability of dormant MTB to convert the pro-drug into an active form is thought to be one of the possible reasons. Employing targeted metabolomics approach, we show that dormant MTB can metabolize INH into its active INH-NAD+ adduct form. Further we show that the dormant bacteria have unaltered gene expression levels of katG and inhA (INH metabolizing enzymes). Transcript levels of drug efflux pump proteins which were low during dormancy did not increase in response to INH treatment. These findings point to an alternative mechanism for INH resistance in dormant MTB, which needs to be further elucidated.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Robitzek EH, Selikoff IJ. Hydrazine derivatives of isonicotinic acid (Rimifon, Marsilid) in the treatment oi active progressive caseous-pneumonic tuberculosis. A preliminary report. Am Rev Tuberc Pulm Dis. 1952;65:402–28.
Jindani A, Aber V, Edwards E, Mitchison D. The early bactericidal activity of drugs in patients with pulmonary tuberculosis. Am Rev Respir Dis. 1980;121:939–49.
Reller LB, Weinstein MP, Woods GL. Susceptibility testing for mycobacteria. Clin Infect Dis. 2000;31:1209–15.
Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591.
Johnsson K, Schultz PG. Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J Am Chem Soc. 1994;116:7425–6.
Lei B, Wei C-J, Tu S-C. Action mechanism of antitubercular isoniazid activation by Mycobacterium tuberculosis KatG, isolation, and characterization of InhA inhibitor. J Biol Chem. 2000;275:2520–6.
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–30.
Takayama K, Wang L, David HL. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1972;2:29–35.
Winder F, Collins P. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. Microbiology. 1970;63:41–8.
Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63.
Rawat R, Whitty A, Tonge PJ. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc Natl Acad Sci. 2003;100:13881–6.
Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102.
Wang F, Jain P, Gulten G, Liu Z, Feng Y, Ganesula K, et al. Mycobacterium tuberculosis dihydrofolate reductase is not a target relevant to the antitubercular activity of isoniazid. Antimicrobial agents and chemotherapy. 2010;54:3776–82.
Zhang Y, Garbe T, Young D. Transformation with katG restores isoniazid‐sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol Microbiol. 1993;8:521–4.
Ferrazoli L, Palaci M, da Silva Telles MA, Ueki SY, Kritski A, Marques LRM, et al. Catalase expression, katG, and MIC of isoniazid for Mycobacterium tuberculosis isolates from Sao Paulo, Brazil. Journal of Infectious Diseases. 1995;171:237–40.
Kapur V, Li L-L, Hamrick MR, Plikaytis BB, Shinnick TM, Telenti A, et al. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Archives of pathology & laboratory medicine. 1995;119:131–8.
Marttila HJ, Soini H, Eerola E, Vyshnevskaya E, Vyshnevskiy BI, Otten TF, et al. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrobial agents and chemotherapy. 1998;42:2443–5.
Telenti A. Genetics of drug resistant tuberculosis. Thorax. 1998;53:793–7.
Gagneux S, Burgos MV, DeRiemer K, Enciso A, Muñoz S, Hopewell PC, et al. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS pathogens. 2006;2:e61.
Yu S, Girotto S, Lee C, Magliozzo RS. Reduced affinity for isoniazid in the S315T mutant of Mycobacterium tuberculosis KatG is a key factor in antibiotic resistance. J Biol Chem. 2003;278:14769–75.
Pym AS, Saint-Joanis B, Cole ST. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun. 2002;70:4955–60.
Kiepiela P, Bishop K, Smith A, Roux L, York D. Genomic mutations in the katG, inhA and aphC genes are useful for the prediction of isoniazid resistance in Mycobacterium tuberculosis isolates from Kwazulu Natal, South Africa. Tuber Lung Dis. 2000;80:47–56.
Vilchèze C, Wang F, Arai M, Hazbón MH, Colangeli R, Kremer L, et al. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nature medicine. 2006;12:1027.
Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–31.
Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–9.
Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chemother. 2007;61:323–31.
Gopinath V, Raghunandanan S, Gomez RL, Jose L, Surendran A, Ramachandran R, et al. Profiling the proteome of Mycobacterium tuberculosis during dormancy and reactivation. Molecular & Cellular Proteomics. 2015;14:2160–76.
Pontino M, Di BG, Fernandez C, Imperiale B, Bodon A, Morcillo N. Evaluation of a colorimetric micromethod for determining the minimal inhibitory concentration of antibiotics against Mycobacterium tuberculosis. Rev Argent Microbiol. 2006;38:145–51.
Louw G, Warren R, van Pittius NG, McEvoy C, Van Helden P, Victor T. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother. 2009;53:3181–9.
Narang A, Giri A, Gupta S, Garima K, Bose M, Varma-Basil M. Contribution of putative efflux pump genes to isoniazid resistance in clinical isolates of Mycobacterium tuberculosis. Int J mycobacteriology. 2017;6:177.
da Silva PEA, Von Groll A, Martin A, Palomino JC. Efflux as a mechanism for drug resistance in Mycobacterium tuberculosis. FEMS Immunol & Med Microbiol. 2011;63:1–9.
Rodrigues L, Villellas C, Bailo R, Viveiros M, Aínsa JA. Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57:751–7.
Li G, Zhang J, Guo Q, Jiang Y, Wei J, Zhao L-l, et al. Efflux pump gene expression in multidrug-resistant Mycobacterium tuberculosis clinical isolates. PLoS ONE. 2015;10:e0119013.
Mahapatra S, Woolhiser LK, Lenaerts AJ, Johnson JL, Eisenach KD, Joloba ML, et al. A novel metabolite of antituberculosis therapy demonstrates host activation of isoniazid and formation of the isoniazid-NAD+ adduct. Antimicrobial agents and chemotherapy. 2012;56:28–35.
Tudó G, Laing K, Mitchison DA, Butcher PD, Waddell SJ. Examining the basis of isoniazid tolerance in nonreplicating Mycobacterium tuberculosis using transcriptional profiling. Future Med Chem. 2010;2:1371–83.
Suter E. Multiplication of tubercle bacilli within phagocytes cultivated in vitro, and effect of streptomycin and isonicotinic acid hydrazide. Am Rev Tuberc Pulm Dis. 1952;65:775–6.
Loots DT. An altered Mycobacterium tuberculosis Metabolome Induced by katG mutations resulting in isoniazid resistance. Antimicrob Agents Chemother. 2014;58:2144–9.
Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol. 2006;62:1220–7.
Choi SW, Maiga M, Maiga MC, Atudorei V, Sharp ZD, Bishai WR, et al. Rapid in vivo detection of isoniazid-sensitive Mycobacterium tuberculosis by breath test. Nature Communications. 2014;5:4989.
Frediani JK, Jones DP, Tukvadze N, Uppal K, Sanikidze E, Kipiani M, et al. Plasma Metabolomics in Human Pulmonary Tuberculosis Disease: A Pilot Study. PLoS ONE. 2014;9:e108854.
Acknowledgements
This study has been funded by the Department of Biotechnology, Government of India [BT/PR5361/MED/29/507/2012 (RAK)]. SR is grateful for the INSPIRE fellowship from the Department of Science and Technology, Government of India. We thank the Mass Spectrometry and Proteomic Core Facility of RGCB for mass spectrometry analysis.
Author contributions
Conceptualization, RAK and SR; methodology, SR, LJ and RAK; investigation, SR and RAK; formal analysis, SR and RAK; writing, SR, LJ and RAK; and resources, RAK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Raghunandanan, S., Jose, L. & Kumar, R.A. Dormant Mycobacterium tuberculosis converts isoniazid to the active drug in a Wayne’s model of dormancy. J Antibiot 71, 939–949 (2018). https://doi.org/10.1038/s41429-018-0098-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41429-018-0098-z
This article is cited by
-
Application of Mycobacterium smegmatis as a surrogate to evaluate drug leads against Mycobacterium tuberculosis
The Journal of Antibiotics (2020)