Abstract
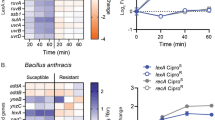
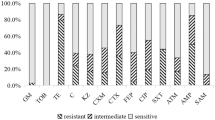
The purpose of this study was focused on the mechanisms of the cross-resistance to tetracycline (TET), piperacillin Sodium (PIP), and gentamicin (GEN) in Staphylococcus aureus (SA) mediated by Rhizoma Coptidis extracts (RCE). The selected strains were exposed continuously to RCE at the sublethal concentrations for 12 days, respectively. The susceptibility change of the drug-exposed strains was determined by analysis of the minimum inhibitory concentration. The 16S rDNA sequencing method was used to identify the RCE-exposed strain. Then the expression of resistant genes in the selected isolates was analyzed by transcriptome sequencing. The results indicated that RCE could trigger the preferential cross-resistance to TET, PIP, and GEN in SA. The correlative resistant genes to the three kinds of antibiotics were upregulated in the RCE-exposed strain, and the mRNA levels of the resistant genes determined by RT-qPCR were consistent with those from the transcriptome analysis. It was suggested from these results that the antibacterial Traditional Chinese Medicines might be a significant factor of causing the bacterial antibiotic-resistance.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
The tables data used to support the findings of this study are included within the article.
References
Tishler PV. Production of penicillin. N Engl J Med. 2005;352:97.
Russell AD. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect Dis. 2003;3:794–803.
Thomas L, Russell AD, Maillard J-Y. Antimicrobial activity of chlorhexidine diacetate and benzalkonium chloride against Pseudomonas aeruginosa and its response to biocide residues. J Appl Microbiol. 2005;98:533–43.
Fernandes P, Ferreira BS, Cabral JM. Solvent tolerance in bacteria: role of efflux pumps and cross-resistance with antibiotics. Int J Antimicrob Agents. 2003;22:211–6.
Thomas L, Russell AD, Maillard J-Y. Antimicrobial activity of chlorhexidine diacetate and benzalkonium chloride against Pseudomonas aeruginosa and its response to biocide residues. J Appl Microbiol. 2005;98:533–43.
Langsrud S, Sidhu MS, Heir E, Askild LH. Bacterial disinfectant resistance a challenge for the food industry. Int Biodeterior Biodegrad. 2003;51:283–90.
Thabit AK, Crandon JL, Nicolau DP. Antimicrobial resistance: impact on clinical and economic outcomes and the need for new antimicrobials. Expert Opin Pharmacother. 2015;16:159–77.
Moremi N, Claus H, Mshana SE. Antimicrobial resistance pattern: a report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infect Dis. 2016;16:756.
Owens RC Jr. Antimicrobial stewardship: concepts and strategies in the 21st century. Diagn Microbiol Infect Dis. 2008;61:110–28.
Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–10. Infect Control Hosp Epidemiol. 2013;34:1–14.
Davin-Regli A, Pagès JM. Cross-resistance between biocides and antimicrobials: an emerging question. Rev Sci Tech. 2012;31:89–104.
Lavilla Lerma L, Benomar N, Casado Muñoz Mdel C, Gálvez A, Abriouel H. Correlation between antibiotic and biocide resistance in mesophilic and psychrotrophic Pseudomonas spp. isolated from slaughterhouse surfaces throughout meat chain production. Food Microbiol. 2015;51:33–44.
Gilbert P, McBain AJ, Bloomfield SF. Biocide abuse and antimicrobial resistance: being clear about the issues. J Antimicrob Chemother. 2002;50:137–9.
Gilbert P, McBain AJ. Biocide usage in the domestic setting and concern about antibacterial and antibiotic resistance. J Infect. 2001;43:85–91.
Lau D, Plotkin BJ. Antimicrobial and biofilm effects of herbs used in traditional Chinese medicine. Nat Prod Commun. 2013;8:1617–20.
Zhang BL, Fan CQ, Dong L, Wang FD, Yue JM. Structural modification of a specific antimicrobial lead against Helicobacter pylori discovered from traditional Chinese medicine and a structure-activity relationship study. Eur J Med Chem. 2010;45:5258–64.
Wong RWK, Hagg U, Samaranayake L, Yuen MKZ, Seneviratne CJ, Kao R. Antimicrobial activity of Chinese medicine herbs against common bacteria in oral biofilm. A pilot study. Int J Oral Maxillofac Surg. 2010;39:599–605.
Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 50-methoxyhydno-carpin, a multidrug pump inhibitor. Proc Natl Acad Sci U S A. 2000;97:1433–7.
Betoni JE, Mantovani RP, Barbosa LN, Stasi LC, Junior AF. Synergism between plant extract and antimicrobial drugs used on Staphylococcus aureus diseases. Mem Inst Oswaldo Cruz. 2006;101:387–90.
Sibanda T, Okoh AI. The challenges of overcoming antibiotic resistance: plant extracts as potential sources of antimicrobial and resistance modifying agents. Afr J Biotechnol. 2007;6:2886–96.
Choi UK, Kim MH, Lee NH. Optimization of antibacterial activity by Gold-Thread (Coptidis Rhizoma Franch) against Streptococcus mutans using evolutionary operation-factorial design technique. J Microbiol Biotechnol. 2007;17:1880–4.
Luo JY, Yan D, Yang MH. Study of the anti-MRSA activity of Rhizoma coptidis by chemical fingerprinting and broth microdilution methods. Chin J Nat Med. 2014;12:393–400.
Wang DW, Liu ZQ, Guo MQ, Liu SY. Structural elucidation and identification of alkaloids in Rhizoma coptidis by electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2004;39:1356–65.
Yokozawa T, Ishida A, Kashiwada Y, Cho EJ, Kim HY, Ikeshiro Y. Coptidis rhizoma: protective effects against peroxynitrite-induced oxidative damage and elucidation of its active components. J Pharm Pharm. 2004;56:547–56.
Asai M, Iwata N, Yoshikawa A, Aizaki Y, Ishiura S, Saido TC, et al. Berberine alters the processing of Alzheimer’s amyloid precursor protein to decrease Abeta secretion. Biochem Biophys Res Commun. 2007;352:498–502.
Wen SQ, Jeyakkumar P, Avula SR, Zhang L, Zhou CH. Discovery of novel berberine imidazoles as safe antimicrobial agents by down regulating ROS generation. Bioorg Med Chem Lett. 2016;26:2768–73.
Wu DM, Lu RC, Chen YQ, Qiu J, Deng CC, Tan Q. Study of cross-resistance mediated by antibiotics, chlorhexidine and Rhizoma coptidis in Staphylococcus aureus. J Glob Antimicrob Resist. 2016;7:61–6.
Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China (Part 2). Beijing, China: China Medical Science Press; 2010, p. 213–4.
Blaszczyk T, Krzyzanowska J, Lamer-Zarawska E. Screening for antimycotic properties of 56 traditional Chinese drugs. Phytother Res. 2000;14:210–2.
Kong WJ, Zhao YL, Xiao XH, Wang JB, Li HB, Li ZL, et al. Spectrum–effect relationships between ultra-performance liquid chromatography fingerprints and antibacterial activities of Rhizoma coptidis. Anal Chim Acta. 2009;634:279–85.
European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. http://www.eucast.org.
Dos Santos HRM, Argolo CS, Argolo-Filho RC, Loguercio LL. A 16S rDNA PCR-based theoretical to actual delta approach on culturable mock communities revealed severe losses of diversity information. BMC Microbiol. 2019;19:74.
Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101.
Qian Y, Linglin G, Teng X, Yun C, Xin Y, Mengmeng H, et al. Amoxicillin administration regimen and resistance mechanisms of staphylococcus aureus established in tissue cage infection model. Front Microbiol. 2019;10:1–9.
Grossman TH. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med. 2016. https://doi.org/10.1101/cshperspect.a025387.
Wencewicz TA. Crossroads of antibiotic resistance and biosynthesis. J Mol Biol. 2019;431:3370–99.
Favrot L, Blanchard JS, Vergnolle O. Bacterial GCN5-related N-acetyltransferases: from resistance to regulation. Biochemistry. 2016;55:989–1002.
Acknowledgements
We are grateful for Professor Xian Zhang for kindly providing the clinical isolates.
Funding
This study was supported by Natural Science Foundation of Guangxi Autonomous Region (NO. 2014GXNSFAA118176), Development and Research Center for China-ASEAN Traditional Medicine and Nanning City Science and Technology Plan [NO. 20131062].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lan, S., Li, Z., Su, A. et al. Study on the mechanisms of the cross-resistance to TET, PIP, and GEN in Staphylococcus aureus mediated by the Rhizoma Coptidis extracts. J Antibiot 74, 330–336 (2021). https://doi.org/10.1038/s41429-021-00407-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41429-021-00407-4