Abstract
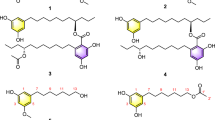
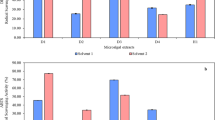
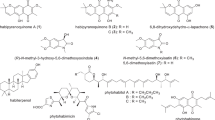
Marine alga-derived fungal strain КММ 4176 was identified as Aspergillus niveoglaucus based on ITS region BenA, CaM and RPB2 gene sequence analysis. The anthraquinone derivatives emodin anthrone (1) and 4-hydroxyemodin anthrone (2), chromone derivative aloesone (3), and indole diketopiperazine alkaloid neoechinulin B (4) were isolated from the ethyl acetate extract of this fungus. In addition, UPLC MS data analysis of the KMM 4176 extract showed the presence of 17 echinulin-family alkaloids, as well as their biogenetic precursor cyclo(l-alanyl-l-tryptophyl) and a number of polyketide compounds. Emodin anthrone and 4-hydroxyemodin anthrone were found as inhibitors of biofilm formation by Staphylococcus aureus with half-maximal inhibitory concentrations (IC50) of 5.5 µM and 23.7 µM, respectively. Moreover, emodin anthrone (1) and 4-hydroxyemodin anthrone (2) inhibited staphylococcal sortase A activity with IC50 of 9.2 µM and 37.6 µM, respectively. Aloesone (3) also inhibited S. aureus biofilm formation but was less active. The first data on neoechinulin B (4) antibiofilm activity and sortase A inhibition were obtained. The positive effects of the isolated compounds on the growth of HaCaT keratinocytes infected with S. aureus were also observed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Calabon MS, Jones EBG, Pang K-L, Abdel-Wahab MA, Jin J, Devadatha B, et al. Updates on the classification and numbers of marine fungi. Bot Mar. 2023;66:213–38.
Hyde KD, Jones EBG, Leaño E, Pointing SB, Poonyth AD, Vrijmoed LLP. Role of fungi in marine ecosystems. Biodivers Conserv. 1998;7:1147–61.
Wijayawardene NN, Dai D-Q, Jayasinghe PK, Gunasekara SS, Nagano Y, Tibpromma S, et al. Ecological and oceanographic perspectives in future marine fungal taxonomy. J Fungi. 2022;8:1141.
Gladfelter AS, James TY, Amend AS. Marine fungi. Curr Biol. 2019;29:R191–R195.
Bilal L, Asaf S, Hamayun M, Gul H, Iqbal A, Ullah I, et al. Plant growth promoting endophytic fungi Asprgillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis. 2018;76:117–27.
El-Bondkly EAM, El-Bondkly AAM, El-Bondkly AAM. Marine endophytic fungal metabolites: a whole new world of pharmaceutical therapy exploration. Heliyon. 2021;7:e06362.
Gao L-W, Zhang P. An update on chemistry and bioactivities of secondary metabolites from the marine algal-derived endophytic fungi. Phytochem Rev. 2023;22:587–614.
Ji N-Y, Wang B-G. Mycochemistry of marine algicolous fungi. Fungal Diversity. 2016;80:301–42.
WHO. Global action plan on antimicrobial resistance. WHO, (2016).
Esposito S, Blasi F, Curtis N, Kaplan S, Lazzarotto T, Meschiari M, et al. New Antibiotics for Staphylococcus aureus Infection: An Update from the World Association of Infectious Diseases and Immunological Disorders (WAidid) and the Italian Society of Anti-Infective Therapy (SITA). Antibiotics. 2023;12:742.
Yurchenko AN, Smetanina OF, Ivanets EV, Phan TTH, Ngo NTD, Zhuravleva OI, et al. Auroglaucin-related neuroprotective compounds from Vietnamese marine sediment-derived fungus Aspergillus niveoglaucus. Nat Prod Res. 2020;34:2589–94.
Smetanina OF, Yurchenko AN, Girich (Ivanets) EV, Trinh PTH, Antonov AS, Dyshlovoy SA, et al. Biologically active echinulin-related indolediketopiperazines from the marine sediment-derived fungus Aspergillus niveoglaucus. Molecules. 2020;25:61.
Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, et al. Identification and nomenclature of the genus Penicillium. Stud Mycol. 2014;78:343–71.
Leshchenko EV, Berdyshev DV, Yurchenko EA, Antonov AS, Borkunov GV, Kirichuk NN, et al. Bioactive polyketides from the natural complex of the sea urchin-associated fungi Penicillium sajarovii KMM 4718 and Aspergillus protuberus KMM 4747. Int J Mol Sci. 2023;24:16568.
Kumar S, Stecher G, Li M, Knyaz C, Tamura KMEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol. 1992;9:678–87.
Nesterenko LE, Popov RS, Zhuravleva OI, Kirichuk NN, Chausova VE, Krasnov KS, et al. A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum. Fermentation. 2023;9:337.
Chambers MC, MacLean B, Burke R, Amodei D, Ruderman DL, Neumann S, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–20.
Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010;11:395.
Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminformatics. 2016;8:3.
Campbell J. High‐throughput assessment of bacterial growth inhibition by optical density measurements. Curr Protoc. Chem Biol. 2010;2:195–208.
Kifer D, Mužinić V, Klarić MŠ. Antimicrobial potency of single and combined mupirocin and monoterpenes, thymol, menthol and 1, 8-cineole against Staphylococcus aureus planktonic and biofilm growth. J Antibiot (Tokyo). 2016;69:689–96.
Grosdidier A, Zoete V. Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–277.
Brooks BR, Brooks Iii CL, Mackerell Jr AD, Nilsson L, Petrella RJ, Roux B, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–614.
Haberthür U, Caflisch A. FACTS: Fast analytical continuum treatment of solvation. J Comput Chem. 2008;29:701–15.
Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011;32:2149–59.
Yurchenko EA, Khmel OO, Nesterenko LE, Aminin DL. The Kelch/Nrf2 antioxidant system as a target for some marine fungal metabolites. Oxygen. 2023;3:374–85.
Abegaz B, Dagne E. Antracene derivatives of Rhamnus prinoides. Bull Chem Soc Ethop. 1988;2:15–20.
Cameron DW, Edmonds JS, Raverty WD. Oxidation of emodin anthrone and stereochemistry of emodin bianthrone. Aust J Chem. 1976;29:1535–48.
Tan JJ, Liu XY, Yang Y, Li FH, Tan CH, Li YM. Aspergillolide, a new 12-membered macrolide from sea cucumber-derived fungus Aspergillus sp. S-3-75. Nat Prod Res. 2020;34:1131–7.
Dossena, A, Marchelli, R, Pochini A. New metabolites of Aspergillus amstelodami related to the biogenesis of neoechinulin. J Chem Soc Chem Commun. 1974:771–2.
Lv D, Xia J, Guan X, Lai Q, Zhang B, Lin J, et al. Indole diketopiperazine alkaloids isolated from the marine-derived fungus Aspergillus chevalieri MCCC M23426. Front Microbiol. 2022;7:950857.
Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12:547–69.
Nitulescu G, Margina D, Zanfirescu A, Olaru OT, Nitulescu GM. Targeting bacterial sortases in search of anti-virulence therapies with low risk of resistance development. Pharmaceuticals. 2021;14:415.
Pishesha N, Ingram JR, Ploegh HL, Sortase A. A model for transpeptidation and its biological applications. Annu Rev Cell Dev Biol. 2018;34:163–88.
Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharm Rev. 2008;60:128–41.
Nitulescu G, Margina D, Zanfirescu A, Olaru OT, Nitulescu GM. Molecular docking and screening studies of new natural sortase A inhibitors. Int J Mol Sci. 2017;18:2217.
El-Kashef DH, Obidake DD, Schiedlauske K, Deipenbrock A, Scharf S, Wang H, et al. Indole diketopiperazine alkaloids from the marine sediment-derived fungus Aspergillus chevalieri against pancreatic ductal adenocarcinoma. Mar Drugs. 2024;22:5.
Chen AJ, Hubka V, Frisvad JC, Visagie CM, Houbraken J, Meijer M, et al. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Stud Mycol. 2017;88:37–135.
Nies J, Li SM. Prenylation and dehydrogenation of a C2-reversely prenylated diketopiperazine as a branching point in the biosynthesis of echinulin family alkaloids in Aspergillus ruber. ACS Chem Biol. 2021;16:185–92.
Radzom M, Zeeck A, Antal N, Fiedler HP. Fogacin, a novel cyclic octaketide produced by Streptomyces strain Tü 6319. J Antibiot. 2006;59:315–7.
Shao C, Wang C, Wei M, Gu Y, Xia X, She Z, et al. Structure elucidation of two new xanthone derivatives from the marine fungus Penicillium sp. (ZZF 32#) from the South China Sea. Magn Reson Chem. 2008;46:1066–9.
Wang C-N, Lu H-M, Gao C-H, Guo L, Zhan Z-Y, Wang J-J, et al. Cytotoxic benzopyranone and xanthone derivatives from a coral symbiotic fungus Cladosporium halotolerans GXIMD 02502. Nat Prod Res. 2021;35:5596–603.
Hao Y-J, Zou Z-B, Xie M-M, Zhang Y, Xu L, Yu H-Y, et al. Ferroptosis inhibitory compounds from the deep-sea-derived fungus Penicillium sp. MCCC 3A00126. Mar Drugs. 2023;21:234.
Das P, Babbar P, Malhotra N, Sharma M, Jachak GR, Gonnade RG, et al. Specific stereoisomeric conformations determine the drug potency of cladosporin scaffold against malarial parasite. J Med Chem. 2018;61:5664–78.
Scott PM, Van Walbeek W, MacLean WM. Cladosporin, a new antifungal metabolite from Cladosporium cladosporioides. J Antibiot. 1971;24:747–55.
Jacyno JM, Harwood JS, Cutler HG, Lee MK. Isocladosporin, a biologically active isomer of cladosporin from Cladosporium cladosporioides. J Nat Prod. 1993;56:1397–401.
Putkaradze N, Dato L, Kırtel O, Hansen J, Welner DH. Enzymatic glycosylation of aloesone performed by plant UDP-dependent glycosyltransferases. Glycobiology. 2024;34:cwae050.
Abe I, Watanabe T, Lou W, Noguchi H. Active site residues governing substrate selectivity and polyketide chain length in aloesone synthase. FEBS J. 2006;273:208–18.
Navarro-Muñoz JC, Collemare J. Evolutionary histories of type III polyketide synthases in fungi. Front Microbiol. 2020;10:3018.
Gessler N, Egorova A, Belozerskaya T. Fungal anthraquinones. Appl Biochem Microbiol. 2013;49:85–99.
Anke H, Kolthoum I, Zähner H, Laatsch H. Metabolic products of microorganisms. 185. The anthraquinones of the Aspergillus glaucus group. I. Occurrence, isolation, identification and antimicrobial activity. Arch Microbiol. 1980;126:223–30.
Kurobane I, Vining LC, Gavin McInnes A. Biosynthetic relationships among the secalonic acids isolation of emodin, endocrocin and secalonic acids from Pyrenochaeta terrestris and Aspergillus aculeatus. J Antibiot. 1979;32:1256–66.
Hafez Ghoran S, Taktaz F, Ayatollahi SA, Kijjoa A. Anthraquinones and their analogues from marine-derived fungi: chemistry and biological activities. Mar Drugs. 2022;20:474.
Ha Y, Zhou Y, Ma M, Wang N, Wang P, Zhang Z. Antimicrobial metabolites from the marine-derived fungus Aspergillus sp. ZZ1861. Phytochemistry. 2024;224:114164.
Chen Z-G, Fujii I, Ebizuka Y, Sankawa U. Purification and characterization of emodinanthrone oxygenase from Aspergillus terreus. Phytochemistry. 1995;38:299–305.
Mondal A, Saha N, Rajput A, Singh SK, Roy B, Husain SM. Chemoenzymatic reduction of citreorosein and its implications on aloe-emodin and rugulosin C (bio)synthesis. Org Biomol Chem. 2019;17:8711–5.
Singh SK, Mondal A, Saha N, Husain SM. Identification and characterization of an anthrol reductase from Talaromyces islandicus (Penicillium islandicum) WF-38-12. Green Chem. 2019;21:6594–9.
Shakour ZT, Farag MA. Diverse host-associated fungal systems as a dynamic source of novel bioactive anthraquinones in drug discovery: Current status and future perspectives. J Adv Res. 2022;39:257–73.
Zhao J, Zhou L, Wang J, Shan T, Zhong L, Liu X, et al. Endophytic fungi for producing bioactive compounds originally from their host plants. Curr Res, Technol Educ Trop Appl Microbiol. Microb Biotechnol. 2010;1:567–76.
Qun T, Zhou T, Hao J, Wang C, Zhang K, Xu J, et al. Antibacterial activities of anthraquinones: structure–activity relationships and action mechanisms. RSC Med Chem. 2023;14:1446–71.
Janeczko M. Emodin reduces the activity of (1, 3)-D-glucan synthase from and does not interact with caspofungin. Pol J Microbiol. 2018;67:463–70.
Lu C, Wang H, Lv W, Xu P, Zhu J, Xie J, et al. Antibacterial properties of anthraquinones extracted from rhubarb against Aeromonas hydrophila. Fish Sci. 2011;77:375–84.
Raghuveer D, Pai VV, Murali TS, Nayak R. Exploring anthraquinones as antibacterial and antifungal agents. ChemistrySelect. 2023;8:e202204537.
Zhuravleva OI, Chingizova EA, Oleinikova GK, Starnovskaya SS, Antonov AS, Kirichuk NN, et al. Anthraquinone derivatives and other aromatic compounds from marine fungus Asteromyces cruciatus KMM 4696 and their effects against Staphylococcus aureus. Mar Drugs. 2023;21:431.
Chalothorn T, Rukachaisirikul V, Phongpaichit S, Pannara S, Tansakul C. Synthesis and antibacterial activity of emodin and its derivatives against methicillin-resistant Staphylococcus aureus. Tetrahedron Lett. 2019;60:151004.
Đukanović S, Ganić T, Lončarević B, Cvetković S, Nikolić B, Tenji D, et al. Elucidating the antibiofilm activity of frangula emodin against Staphylococcus aureus biofilms. J Appl Microbiol. 2022;132:1840–55.
Lu F, Wu X, Hu H, He Z, Sun J, Zhang J, et al. Emodin combined with multiple-low-frequency, low-intensity ultrasound to relieve osteomyelitis through sonoantimicrobial chemotherapy. Microbiol Spectr. 2022;10:e00544–00522.
Wang Y, Xiong Z, Li C, Liu D, Li X, Xu J, et al. Multiple Beneficial Effects of Aloesone from Aloe vera on LPS-Induced RAW264.7 Cells, Including the Inhibition of Oxidative Stress, Inflammation, M1 Polarization, and Apoptosis. Molecules. 2023;28:1617.
Kim K-S, Cui X, Lee D-S, Sohn JH, Yim JH, Kim Y-C, et al. Anti-Inflammatory Effect of Neoechinulin A from the Marine Fungus Eurotium sp. SF-5989 through the Suppression of NF-кB and p38 MAPK Pathways in Lipopolysaccharide-Stimulated RAW264.7 Macrophages. Molecules. 2013;18:13245–59.
Yan L-H, Li P-H, Li X-M, Yang S-Q, Liu K-C, Wang B-G, et al. Chevalinulins A and B, Proangiogenic Alkaloids with a Spiro[bicyclo[2.2.2]octane-diketopiperazine] Skeleton from Deep-Sea Cold-Seep-Derived Fungus Aspergillus chevalieri CS-122. Org Lett. 2022;24:2684–8.
Sharifi-Rad J, Bahukhandi A, Dhyani P, Sati P, Capanoglu E, Docea AO, et al. Therapeutic potential of neoechinulins and their derivatives: an overview of the molecular mechanisms behind pharmacological activities. Frontiers in Nutrition. 2021;8:664197.
Yan L-H, Du F-Y, Li X-M, Yang S-Q, Wang B-G, Li X. Antibacterial indole diketopiperazine alkaloids from the deep-sea cold seep-derived fungus Aspergillus chevalieri. Mar Drugs. 2023;21:195.
Song W, Wang L, Zhao Y, Lanzi G, Wang X, Zhang C, et al. Hibifolin, a natural sortase a inhibitor, attenuates the pathogenicity of Staphylococcus aureus and enhances the antibacterial activity of cefotaxime. Microbiol Spectrum. 2022;10:e00950–00922.
Jiang T, Yuan D, Wang R, Zhao C, Xu Y, Liu Y, et al. Echinacoside, a promising sortase A inhibitor, combined with vancomycin against murine models of MRSA-induced pneumonia. Med Microbiol Immunol. 2023;212:421–35.
Otto M. Critical assessment of the prospects of quorum-quenching therapy for Staphylococcus aureus infection. Int J Mol Sci. 2023;24:4025.
Bapat RA, Parolia A, Chaubal T, Yang HJ, Kesharwani P, Phaik KS, et al. Recent update on applications of quaternary ammonium silane as an antibacterial biomaterial: a novel drug delivery approach in dentistry. Frontiers in Microbiology. 2022;13:927282.
Butler MS, Henderson IR, Capon RJ, Blaskovich MAT. Antibiotics in the clinical pipeline as of December 2022. J Antibiot (Tokyo). 2023;76:431–73.
Acknowledgements
This study was carried out using the equipment of the Collective Facilities Center “The Far Eastern Center for Structural Molecular Research (NMR/MS) PIBOC FEB RAS” and using the Collective Facilities Center “Collection of Marine Microorganisms PIBOC FEB RAS”. The authors thank Technoinfo company (https://technoinfo.ru) and personally Alexander Smirnov for the opportunity to use the real-time cell imager JuLiStage for this research.
Funding
This research was funded by a grant from the Ministry of Science and Higher Education of Russian Federation 15.BRK.21.0004 (Contract No.: 075-15-2021-1052).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Materials: The following supporting information can be downloaded at:
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shlyk, N.P., Yurchenko, E.A., Leshchenko, E.V. et al. The secondary metabolites of the alga-derived fungus Aspergillus niveoglaucus КММ 4176 and their antimicrobial and antibiofilm activities. J Antibiot 78, 314–329 (2025). https://doi.org/10.1038/s41429-025-00811-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41429-025-00811-0