Abstract
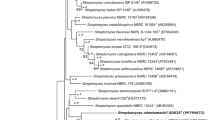
A novel actinobacterium strain cg36T, from the rhizosphere of Cyclosorus parasiticus, was subjected to a polyphasic taxonomic study. 16S rRNA gene sequence analysis showed that strain cg36T shared the highest 16S rRNA gene sequence similarity to Streptomyces lavendofoliae NBRC 12882T. Phylogenetic analysis of 16S rRNA gene sequence indicated that strain cg36T formed a distinct branch within the genus Streptomyces. Phylogenetic analysis of five housekeeping gene and whole genome sequences showed that strain cg36T was closely related to Streptomyces crystallinus JCM 5067T and Streptomyces noboritoensis JCM 4557T. But, overall genome related index (OGRI) and multilocus sequence analysis (MLSA) confirmed that strain cg36T was clearly different from them. A comparison of differential features among strain cg36T, S. crystallinus CGMCC 4.1600T and S. noboritoensis CGMCC 4.1457T provided, at least to some extent, some useful information for strain cg36T as an independent species. In addition, OGRI values and MLSA evolutionary distance between strain cg36T and other strains, which had 16S rRNA gene sequence similarity of >98.65% to strain cg36T, further demonstrated that strain cg36T was a new species. Whole-cell hydrolysates of strain cg36T contained ll-diaminopimelic acid and whole-cell sugars contain glucose. The predominant cellular fatty acids (>10%) were anteiso-C15:0, iso-C15:0 and C16:0. The DNA G + C content of the genome sequence, consisting of 9,022,416 bp, was 72.5%. All these data indicated that strain cg36T represents a novel Streptomyces species, for which the name Streptomyces cyclosori sp. nov. is proposed. The type strain is strain cg36T (=MCCC 1K09286T = JCM 37520T).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet. 2024;404:1199–226.
Genilloud O. The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek. 2014;106:173–88.
Rossiter SE, Fletcher MH, Wuest WM. Natural products as platforms to overcome antibiotic resistance. Chem Rev. 2017;117:12415–74.
Moloney MG. Natural products as a source for novel antibiotics. Trends Pharm Sci. 2016;37:689–701.
Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol Res. 2014;169:262–78.
Waksman SA, Henrici AT. The nomenclature and classification of the Actinomycetes. J Bacteriol. 1943;46:337–41.
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70:5607–12.
van Bergeijk DA, Terlouw BR, Medema MH, van Wezel GP. Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat Rev Microbiol. 2020;18:546–58.
Ramarajan M, Devilla R, Dow L, Walsh N, Mead O, Zakeel MCM, Gallart M, Richardson AE, Thatcher LF. Genomic and untargeted metabolomic analysis of secondary metabolites in the Streptomyces griseoaurantiacus strain MH191 shows media-based dependency for the production of bioactive compounds with potential antifungal activity. J Agric Food Chem. 2024;72:24432–48.
Sriragavi G, Sangeetha M, Santhakumar M, Lokesh E, Nithyalakshmi M, Saleel CA, Balagurunathan R. Exploring antibacterial properties of bioactive compounds isolated from Streptomyces sp. in bamboo rhizosphere soil. ACS Omega. 2023;8:36333–43.
Zhou B, Hu ZJ, Zhang HJ, Li JQ, Ding WJ, Ma ZJ. Bioactive staurosporine derivatives from the Streptomyces sp. NB-A13. Bioorganic Chem. 2019;82:33–40.
Gencbay T, Saygin H, Guven K, Topkara AR, Saricaoglu S, Sahin N, Isik K. Streptomyces scabichelini sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2021;71:004639.
Atlas RM. Handbook of microbiological media. Boca Raton: CRC Press; 1993.
Jiang CR, Ruan JS. Two new species and a new variety of Ampullarella. Acta Microbiol Sin. 1982;22:207–11.
Shirling EB, Gottlieb D. Methods for characterisation of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Ridgway R. Color standards and color nomenclature. Washington, DC. 1912. pp.1–43.
Saimee Y, Duangmal K. Streptomyces spirodelae sp. nov., isolated from duckweed. Int J Syst Evol Microbiol. 2021;71:005106.
Xu LH, Li WJ, Liu ZH, Jiang CL. Actinomycete systematics: principle, methods and practice. Beijing: Science Press; 2007.
Hasegawa T, Takizawa M, Tanida S. A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol. 1983;29:319–22.
Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:435–43.
Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol. 1996;46:1088–92.
Mo P, Li K, Zhou J, Zhou F, He J, Zou WS, Gao J. Nocardiopsis changdeensis sp. nov., an endophytic actinomycete isolated from the roots of Eucommia ulmoides Oliv. J Antibiot. 2023;76:191–7.
Chalita M, Kim YO, Park S, Oh HS, Cho JH, Moon J, Baek N, Moon C, Lee K, Yang J, et al. EzBioCloud: a genome-driven database and platform for microbiome identification and discovery. Int J Syst Evol Microbiol. 2024;74:006421.
Rong X, Huang Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA–DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst Appl Microbiol. 2012;35:7–18.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Rzhetsky A, Nei M. A simple method for estimating and testing minimum evolution trees. Mol Biol Evol. 1992;9:945–67.
Kumar S, Stecher G, Suleski M, Sanderford M, Sharma S, Tamura K. MEGA12: Molecular Evolutionary Genetics Analysis version 12 for adaptive and green computing. Mol Biol Evol. 2024;41:1–9.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42:D206–14.
Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, Fetter A, Terlouw BR, Metcalf WW, Helfrich EJN, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualization. Nucleic Acid Res. 2023;51:W46–50.
Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acid Res. 2023;5:D690–9.
Meier-Kolthoff JP, Sardà Carbasse J, Peinado-Olarte RL, Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022;50:D801–7.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Meier-Kolthof JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinforma. 2013;14:60.
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31.
Vandamme P, Sutcliffe I. Out with the old and in with the new: time to rethink twentieth century chemotaxonomic practices in bacterial taxonomy. Int J Syst Evol Microbiol. 2021;71:005127.
Hu SR, Li KQ, Zhang YF, Wang YF, Li Fu, Xiao Y, Tang XK, Gao J. New insights into the threshold values of multilocus sequence analysis, average nucleotide identity and digital DNA–DNA hybridization in delineating Streptomyces species. Front Microbiol. 2022;13:910277.
Auch AF, Jan MV, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom Sci. 2010;2:117–34.
Riesco R, Carro L, Román-Ponce B, Prieto C, Blom J, et al. Defining the species Micromonospora saelicesensis and Micromonospora noduli under the framework of genomics. Front Microbiol. 2018;9:1360.
Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–5.
Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, Ju KS, et al. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 2017;110:563–83.
Xie JJ, Zhang HY, Xu X, Yang KX, Ou JC, Yang DF, Jiang Y, Jiang MG, Shen NK. Streptomyces fuscus sp. nov., a brown-black pigment producing actinomycete isolated from dry mudflat sand. Int J Syst Evol Microbiol. 2023;73:0.006047.
Qi DF, Liu Q, Zou LP, Zhang MY, Li K, Zhao YK, Chen YF, Feng JT, Zhou DB, Wei YZ, Wang W, Zhang L, Xie JH. Taxonomic identification and antagonistic activity of Streptomyces luomodiensis sp. nov. against phytopathogenic fungi. Front Microbiol. 2024;15:1402653 27.
Acknowledgements
We would like to express appreciation to Prof. Aharon Oren (Department of Plant and Environmental Sciences and the Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem) for his assistance with the etymology check.
Funding
This work was supported by the Hunan Provincial Natural Science Foundation of China and Xiangtan Science and Technology Bureau (2022JJ50125).
Author information
Authors and Affiliations
Contributions
RJG and YZC conducted experiments, prepared figures and tables. RJG and YX purchased type strains. RJG and YZC wrote the main manuscript text. JG corrected and reviewed the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, R., Chen, Y., Xiao, Y. et al. Streptomyces cyclosori sp. nov., a novel actinobacterium from the rhizosphere soil of Cyclosorus parasiticus (L.) Farw. J Antibiot 78, 666–673 (2025). https://doi.org/10.1038/s41429-025-00857-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41429-025-00857-0