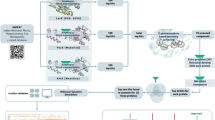
Abstract
Antibiotic resistance has emerged as a critical global public health challenge. Quorum sensing (QS), a density-dependent regulatory mechanism, plays a pivotal role in bacterial pathogenesis by coordinating virulence factor expression, making it a critical target for antivirulence therapy. Leveraging a drug repositioning strategy, this study investigated the antivirulence potential of drugs in the database of DrugBank on the common opportunistic pathogen Pseudomonas aeruginosa by virtual screening. Molecular docking analysis predicted that the antitumor drug, Tirazone, could bind to the core QS regulatory proteins, LasR, RhlR, and PqsR of P. aeruginosa with abundant active sites, whereas the binding free energies were higher than those of the native QS signals. In vitro experiments demonstrated that Tirazone significantly suppressed virulence factor secretion, cell motilities, and biofilm formation in the model P. aeruginosa strain PAO1, and downregulated the expression of a series of QS-related genes with low effective concentration (≤ 8 μM). A competitive binding model of QS signal molecules further elucidated that Tirazone interfered with QS signaling by competitively inhibiting the function of LasR, RhlR, and PqsR. Additionally, Tirazone treatment significantly protected Caenorhabditis elegans and mouse models from P. aeruginosa infection, and reduced the bacterial loads and pathological lesions in mouse lungs. Moreover, Tirazone demonstrated synergistic effects with polymyxin B, levofloxacin, and amikacin, significantly enhancing their bactericidal efficacy in treating P. aeruginosa. This study reveals the molecular mechanism underlying Tirazone’s multi-target intervention in the QS system, and provides an experimental foundation for developing combination therapies based on antivirulence strategies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others

Data availability
The datasets supporting the findings of this study are publicly available in the Figshare repository and can be accessed via the permanent https://doi.org/10.6084/m9.figshare.29301779.
References
Hutchings MI, Truman AW, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51:72–80.
Oliveira M, et al. An overview of the recent advances in antimicrobial resistance. Microorganisms. 2024;12:1920.
Naghavi M, et al. Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet. 2024;404:1199–226.
Muteeb G, Rehman MT, Shahwan M, Aatif M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: a narrative review. Pharmaceuticals. 2023;16:1615.
Li J, Jia T, Yang L. Targeting anti-virulence factor strategies of bacterial pathogens. Biosaf Health. 2025;7:1–4.
Xiao Y, et al. Impact of quorum-sensing signaling molecules in Gram-negative bacteria on host cells: current understanding and future perspectives. Gut Microbes. 2022;14:2039048.
Azimi S, Klementiev AD, Whiteley M, Diggle SP. Bacterial quorum sensing during infection. Annu Rev Microbiol. 2020;74:201–19.
Inoue Y, Togashi N, Hamashima H. Farnesol-induced disruption of the Staphylococcus aureus cytoplasmic membrane. Biol Pharm Bull. 2016;39:653–6.
Naga NG, Shaaban MI, El-Metwally MM. An insight on the powerful of bacterial quorum sensing inhibition. Eur J Clin Microbiol Infect Dis. 2024;43:2071–81.
Qin S, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Sig Transduct Target Ther. 2022;7:199.
Chadha J, Harjai K, Chhibber S. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ Microbiol. 2022;24:2630–56.
Lee J, Zhang L. The hierarchy quorum-sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41.
Nadal Jimenez P, et al. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76:46–65.
Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427.
Papenfort K, Bassler BL. Quorum-sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–88.
Zhao K. Pseudomonas aeruginosa quorum-sensing and type VI secretion system can direct interspecific coexistence during evolution. Front Microbiol. 2018;9:2287.
Yuan Y, et al. Repurposing dimetridazole and ribavirin to disarm Pseudomonas aeruginosa virulence by targeting the quorum-sensing system. Front Microbiol. 2022;13:978502.
Krishnan T, Yin W-F, Chan K-G. Inhibition of quorum-sensing-controlled virulence factor production in Pseudomonas aeruginosa PAO1 by ayurveda spice clove (Syzygium Aromaticum) bud extract. Sensors. 2012;12:4016–30.
Du L, et al. Dockey: a modern integrated tool for large-scale molecular docking and virtual screening. Brief Bioinform. 2023;24:bbad047.
Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–75.
Toussaint, J-P et al. Gene duplication in Pseudomonas aeruginosa improves growth on adenosine. J Bacteriol 2017;199:e00261–17.
Zhao K, et al. Genetic and functional diversity of Pseudomonas aeruginosa in patients with chronic obstructive pulmonary disease. Front Microbiol. 2020;11:598478.
Das T, Manefield M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE. 2012;7:e46718.
Wang S, et al. Inhibition of virulence factors and biofilm formation by wogonin attenuates pathogenicity of Pseudomonas aeruginosa PAO1 via targeting pqs quorum-sensing system. Int J Mol Sci. 2021;22:12699.
Shukla, SK, Rao, TS. An improved crystal violet assay for biofilm quantification in 96-well microtitre plate. 2017 Preprint at https://doi.org/10.1101/100214.
Pfeifer V, Beier S, Alirezaeizanjani Z, Beta C. Role of the two flagellar stators in swimming motility of Pseudomonas putida. mBio. 2022;13:e02182–22.
Ha D–G, Kuchma, SL, O’Toole, GA. Plate-Based Assay for Swarming Motility. In: Filloux A, & Ramos J-L, editors. Pseudomonas aeruginosa: Methods and Protocols. New York: Springer-Verlag; 2014. p. 67–72.
Wen F, et al. Discovery of psoralen as a quorum-sensing inhibitor suppresses Pseudomonas aeruginosa virulence. Appl Microbiol Biotechnol. 2024;108:222.
Wu Y, et al. Multifaceted quorum-sensing inhibiting activity of 3-(Benzo[d] [1,3] dioxol-4-yl) oxazolidin-2-one mitigates Pseudomonas aeruginosa virulence. Virulence. 2025;16:2479103.
Huang T, et al. A potent antibiotic combination of linezolid and polymycin B nonapeptide against Klebsiella pneumoniae infection in vitro and in vivo. Front Pharmacol. 2022;13:887941.
Liu Q, Yin X, Languino LR, Altieri DC. Evaluation of drug combination effect using a Bliss independence dose–response surface model. Stat Biopharm Res. 2018;10:112–22.
Desalermos A, Muhammed M, Glavis-Bloom J, Mylonakis E. Using Caenorhabditis elegans for antimicrobial drug discovery. Expert Opin Drug Discov. 2011;6:645–52.
Mazzolini R, et al. Engineered live bacteria suppress Pseudomonas aeruginosa infection in mouse lung and dissolve endotracheal-tube biofilms. Nat Biotechnol. 2023;41:1089–98.
Muhlen, S, Dersch, P. Anti-virulence strategies to target bacterial infections. In: Stadler M, & Dersch P, editors. How to overcome the antibiotic crisis. Cham: Springer International Publishing; 2015. p. 147–83.
Alatraktchi FA, Svendsen WE, Molin S. Electrochemical detection of pyocyanin as a biomarker for Pseudomonas aeruginosa: a focused review. Sensors. 2020;20:5218.
Carradori S, et al. Biofilm and quorum-sensing inhibitors: the road so far. Expert Opin Ther Pat. 2020;30:917–30.
Martínez E, Campos-Gómez J. Oxylipins produced by Pseudomonas aeruginosa promote biofilm formation and virulence. Nat Commun. 2016;7:13823.
Sandri A, et al. Inhibition of Pseudomonas aeruginosa secreted virulence factors reduces lung inflammation in CF mice. Virulence. 2018;9:1008–18.
Conery AL, Larkins-Ford J, Ausubel FM, Kirienko NV. High-throughput screening for novel anti-infectives using a C. elegans pathogenesis model. CP Chem Biol. 2014;6:25–37.
Marcu L, Olver I. Tirapazamine: from bench to clinical trials. Curr Clin Pharmacol. 2006;1:71–9.
Gatzemeier U, et al. Tirapazamine-cisplatin: the synergy. Br J Cancer. 1998;77:15–7.
Adam M, et al. Tirapazamine plus cisplatin and irradiation in a mouse model: improved tumor control at the cost of increased toxicity. J Cancer Res Clin Oncol. 2007;134:137–46.
Adam M, et al. Untersuchungen zur Toxizität von Tirapazamin plus Cisplatin in einem Maus-Tumormodell. Strahlenther Onkol. 2006;182:231–9.
Okeke IN, et al. The scope of the antimicrobial resistance challenge. Lancet. 2024;403:2426–38.
Luo Q, et al. ESKAPE in China: epidemiology and characteristics of antibiotic resistance. Emerg Microbes Infect. 2024;13:2317915.
Monroy-Pérez E, et al. Molecular properties of virulence and antibiotic resistance of Pseudomonas aeruginosa causing clinically critical infections. Pathogens. 2024;13:868.
Hurley MN, Cámara M, Smyth AR. Novel approaches to the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. Eur Respir J. 2012;40:1014–23.
Yuan M, et al. Repurposing the anticancer drug cisplatin with the aim of developing novel Pseudomonas aeruginosa infection control agents. Beilstein J Org Chem. 2018;14:3059–69.
Imperi F, et al. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc Natl Acad Sci USA. 2013;110:7458–63.
Hegazy WAH, et al. Repurposing anti-diabetic drugs to cripple quorum sensing in Pseudomonas aeruginosa. Microorganisms. 2020;8:1285.
Choi H-Y, Le DD, Kim W-G. Curvularin isolated from Phoma macrostoma is an antagonist of RhlR quorum sensing in Pseudomonas aeruginosa. Front Microbiol. 2022;13:913882.
Ding F, et al. Multidrug-resistant Pseudomonas aeruginosa is predisposed to lasR mutation through up-regulated activity of efflux pumps in non-cystic fibrosis bronchiectasis patients. Front Cell Infect Microbiol. 2022;12:934439.
Soto-Aceves MP, et al. Inactivation of the quorum-sensing transcriptional regulators LasR or RhlR does not suppress the expression of virulence factors and the virulence of Pseudomonas aeruginosa PAO1. Microbiology. 2019;165:425–32.
Deng J, et al. Isovanillin decreases the virulence regulated by the quorum-sensing system of Pseudomonas aeruginosa. Microb Pathog. 2024;196:107010.
Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol. 2006;296:149–61.
Pan AC, Borhani DW, Dror RO, Shaw DE. Molecular determinants of drug–receptor binding kinetics. Drug Discov Today. 2013;18:667–73.
Copeland RA. The drug–target residence time model: a 10-year retrospective. Nat Rev Drug Discov. 2016;15:87–95.
Segala E, et al. Controlling the dissociation of ligands from the adenosine A2A receptor through modulation of salt bridge strength. J Med Chem. 2016;59:6470–9.
Zhou J-W, et al. Attenuation of Pseudomonas aeruginosa biofilm by hordenine: a combinatorial study with aminoglycoside antibiotics. Appl Microbiol Biotechnol. 2018;102:9745–58.
Babylon Education Directorate, Ministry of Education, Iraq et al. Kinetic and thermodynamic principles underlying drug–target interactions: a review article. Int J Med Sci Dent Health. 2025;11:66–70.
Wu Z, et al. New insights into the antimicrobial action and protective therapeutic effect of tirapazamine towards Escherichia coli-infected mice. Int J Antimicrob Agents. 2023;62:106923.
Chu X, Yang Q. Regulatory mechanisms and physiological impacts of quorum sensing in gram-negative bacteria. Infect Drug Resist. 2024;ume 17:5395–410.
Masuda N, et al. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2000;44:3322–7.
Zahedi Bialvaei A, et al. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb Pathog. 2021;153:104789.
Singh M, Sykes EME, Li Y, Kumar A. MexXY RND pump of Pseudomonas aeruginosa PA7 effluxes bi-anionic β-lactams carbenicillin and sulbenicillin when it partners with the outer membrane factor OprA but not with OprM. Microbiology. 2020;166:1095–106.
Seupt A, Schniederjans M, Tomasch J, Häussler S. Expression of the MexXY aminoglycoside efflux pump and presence of an aminoglycoside-modifying enzyme in clinical Pseudomonas aeruginosa isolates are highly correlated. Antimicrob Agents Chemother. 2020;65:e01166–20.
Mlynarcik P, Kolar M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2019;163:28–38.
Lorusso AB, Carrara JA, Barroso CDN, Tuon FF, Faoro H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. IJMS. 2022;23:15779.
Wang J, et al. Research progress on the combination of quorum-sensing inhibitors and antibiotics against bacterial resistance. Molecules. 2024;29:1674.
Acknowledgements
We sincerely thank Prof. Xikun Zhou from Sichuan University for his kind help in coordinating the animal experiments of this study.
Funding
This work was supported by the National Natural Science Foundation of China (32270121 to KZ, 32200525 to L Du and 32400477 to L Dan), the Natural Science Foundation of Sichuan Province (2023NSFSC1231 to L Dan) and the High-level Talent Training Program of Chengdu University (2081920066 to KZ).
Author information
Authors and Affiliations
Contributions
KZ and LDu conceptualized and led the project. MF, XH, YW, SG, QR and YY performed the experiments. XW, L Dan, YC, XZ, and TH facilitated the procurement of essential materials and equipment. MF and XW analyzed the data, interpreted the results, and drafted the thesis. KZ and L Du reviewed and revised the manuscript in its entirety and provided final approval for publication. All authors have reviewed the final manuscript and consented to assume responsibility for all aspects of the research.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, M., Wu, X., Hu, X. et al. Repurposing Tirazone as an effective quorum-sensing inhibitor against Pseudomonas aeruginosa virulence and biofilm formation. J Antibiot (2026). https://doi.org/10.1038/s41429-026-00901-7
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41429-026-00901-7