Abstract
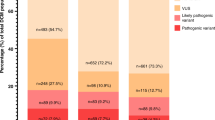
In inherited primary arrhythmia syndromes (PAS) and cardiomyopathies (CMP), the yield of genetic testing varies between 20 and 75% in different diseases according to studies performed in the pre next-generation sequencing (NGS) era. It is unknown whether retesting historical negative samples with NGS techniques is worthwhile. Therefore, we assessed the value of NGS-based panel testing in previously genotype negative–phenotype positive probands. We selected 107 patients (47 PAS and 60 CMP) with a clear phenotype who remained genotype negative after genetic analysis of the main genes implicated in their specific phenotype. Targeted sequencing of the coding regions of 71 PAS- and CMP-related genes was performed. Variant interpretation and classification was done according to a cardiology-specific scoring algorithm (‘Amsterdam criteria’) and the ACMG-AMP criteria. Co-segregation analysis was performed when DNA and clinical data of family members were available. Finally, a genetic diagnosis could be established in 21 patients (20%), 5 PAS (11%) and 16 CMP (27%) patients, respectively. The increased detection rate was due to sequencing of novel genes in 52% of the cases and due to technical failures with the historical analysis in 48%. A total of 118 individuals were informed about their carrier state and either reassured or scheduled for proper follow-up. To conclude, genetic retesting in clinically overt PAS and CMP cases, who were genotype negative with older techniques, resulted in an additional genetic diagnosis in up to 20% of the cases. This clearly supports a policy for genetic retesting with NGS-based panels.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace. 2011;13:1077–1109.
Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46.
Kapplinger JD, Tester DJ, Salisbury BA, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–1303.
Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232.
Koopmann TT, Alders M, Jongbloed RJ, et al. Long QT syndrome caused by a large duplication in the KCNH2 (HERG) gene undetectable by current polymerase chain reaction-based exon-scanning methodologies. Heart Rhythm. 2006;3:52–55.
Eddy CA, MacCormick JM, Chung SK, et al. Identification of large gene deletions and duplications in KCNQ1 and KCNH2 in patients with long QT syndrome. Heart Rhythm. 2008;5:1275–1281.
Tester DJ, Benton AJ, Train L, Deal B, Baudhuin LM, Ackerman MJ. Prevalence and spectrum of large deletions or duplications in the major long QT syndrome-susceptibility genes and implications for long QT syndrome genetic testing. Am J Cardiol. 2010;106:1124–1128.
Barc J, Briec F, Schmitt S, et al. Screening for copy number variation in genes associated with the long QT syndrome: clinical relevance. J Am Coll Cardiol. 2011;57:40–47.
Priori SG, Wilde AA, Horie M, et al. Executive summary: HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Europace. 2013;15:1389–1406.
Elliott PM, Anastasakis A, Borger MA, et al. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J. 2014;35:2733–2779.
Pinto YM, Elliott PM, Arbustini E, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37:1850–1858.
Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532.
van Spaendonck-Zwarts KY, van Rijsingen IA, van den Berg MP, et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years' experience. Eur J Heart Fail. 2013;15:628–636.
Hofman N, Tan HL, Alders M, et al. Yield of molecular and clinical testing for arrhythmia syndromes: report of 15 years' experience. Circulation. 2013;128:1513–1521.
Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP), Seattle, WA. http://evs.gs.washington.edu/EVS/). Accessed February 2017.
Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR, and Consortium GP. A global reference for human genetic variation. Nature. 2015;526:68–74
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291.
Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424.
Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG-AMP Variant-Interpretation Guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;99:247.
Green RC, Goddard KA, Jarvik GP, et al. Clinical Sequencing Exploratory Research Consortium: accelerating evidence-based practice of genomic medicine. Am J Hum Genet. 2016;99:246.
Jarvik GP, Browning BL. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am J Hum Genet. 2016;98:1077–1081.
Fokstuen S, Munoz A, Melacini P, et al. Rapid detection of genetic variants in hypertrophic cardiomyopathy by custom DNA resequencing array in clinical practice. J Med Genet. 2011;48:572–576.
Medlock MM, Tester DJ, Will ML, Bos JM, Ackerman MJ. Repeat long QT syndrome genetic testing of phenotype-positive cases: prevalence and etiology of detection misses. Heart Rhythm. 2012;9:1977–1982.
Crotti L, Marcou CA, Tester DJ, et al. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60:1410–1418.
Robyns T, Kuiperi C, Willems R, Corveleyn A, Nuyens D. Targeted capture sequencing in a large LQTS family reveals a new pathogenic mutation c.2038delG in KCNH2 initially missed due to allelic dropout. Acta Cardiol. 2015;70:747–749.
Tsuji-Wakisaka K, Akao M, Ishii TM, et al. Identification and functional characterization of KCNQ1 mutations around the exon 7-intron 7 junction affecting the splicing process. Biochim Biophys Acta. 2011;1812:1452–1459.
Lin J, Zheng DD, Tao Q, et al. Two novel mutations of the MYBPC3 gene identified in Chinese families with hypertrophic cardiomyopathy. Can J Cardiol. 2010;26:518–522.
Millat G, Lafont E, Nony S, et al. Functional characterization of putative novel splicing mutations in the cardiomyopathy-causing genes. DNA Cell Biol. 2015;34:489–496.
Matthijs G, Souche E, Alders M, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016;24:2–5.
Das KJ, Ingles J, Bagnall RD, Semsarian C. Determining pathogenicity of genetic variants in hypertrophic cardiomyopathy: importance of periodic reassessment. Genet Med. 2014;16:286–293.
Bhuiyan ZA, van den Berg MP, van Tintelen JP, et al. Expanding spectrum of human RYR2-related disease: new electrocardiographic, structural, and genetic features. Circulation. 2007;116:1569–1576.
Ohno S, Omura M, Kawamura M, et al. Exon 3 deletion of RYR2 encoding cardiac ryanodine receptor is associated with left ventricular non-compaction. Europace. 2014;16:1646–1654.
Giudicessi JR, Kapplinger JD, Tester DJ, et al. Phylogenetic and physicochemical analyses enhance the classification of rare nonsynonymous single nucleotide variants in type 1 and 2 long-QT syndrome. Circ Cardiovasc Genet. 2012;5:519–528.
Kapplinger JD, Giudicessi JR, Ye D, et al. Enhanced classification of Brugada syndrome-associated and long-QT syndrome-associated genetic variants in the SCN5A-encoded Na(v)1.5 cardiac sodium channel. Circ Cardiovasc Genet. 2015;8:582–595.
Akinrinade O, Alastalo TP, Koskenvuo JW. Relevance of truncating titin mutations in dilated cardiomyopathy. Clin Genet. 2016;90:49–54.
Kayvanpour E, Sedaghat-Hamedani F, Amr A, et al. Genotype-phenotype associations in dilated cardiomyopathy: meta-analysis on more than 8000 individuals. Clin Res Cardiol. 2017;106:127–139.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Robyns, T., Kuiperi, C., Breckpot, J. et al. Repeat genetic testing with targeted capture sequencing in primary arrhythmia syndrome and cardiomyopathy. Eur J Hum Genet 25, 1313–1323 (2017). https://doi.org/10.1038/s41431-017-0004-3
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-017-0004-3
This article is cited by
-
Patients with unexplained mismatch repair deficiency are interested in updated genetic testing
Hereditary Cancer in Clinical Practice (2020)
-
Identification and Functional Characterization of an ISL1 Mutation Predisposing to Dilated Cardiomyopathy
Journal of Cardiovascular Translational Research (2019)