Abstract
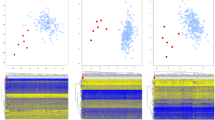
Kabuki syndrome is a monogenic disorder caused by loss of function variants in either of two genes encoding histone-modifying enzymes. We performed targeted sequencing in a cohort of 27 probands with a clinical diagnosis of Kabuki syndrome. Of these, 12 had causative variants in the two known Kabuki syndrome genes. In 2, we identified presumptive loss of function de novo variants in KMT2A (missense and splice site variants), a gene that encodes another histone modifying enzyme previously exclusively associated with Wiedermann-Steiner syndrome. Although Kabuki syndrome is a disorder of histone modification, we also find alterations in DNA methylation among individuals with a Kabuki syndrome diagnosis relative to matched normal controls, regardless of whether they carry a variant in KMT2A or KMT2D or not. Furthermore, we observed characteristic global abnormalities of DNA methylation that distinguished patients with a loss of function variant in KMT2D or missense or splice site variants in either KMT2D or KMT2A from normal controls. Our results provide new insights into the relationship of genotype to epigenotype and phenotype and indicate cross-talk between histone and DNA methylation machineries exposed by inborn errors of the epigenetic apparatus.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3.
Miyake N, Mizuno S, Okamoto N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–10.
Lindsley AW, Saal HM, Burrow TA, et al. Defects of B-cell terminal differentiation in patients with type-1 Kabuki syndrome. J Allerg Clin Immunol. 2016;137:179–87.
Bögershausen N, Gatinois V, Riehmer V, et al. Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-Linked Kabuki syndrome subtype 2. Hum Mutat. 2016;37:847–64.
Bjornsson HT, Benjamin JS, Zhang L, et al. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci Transl Med. 2014;6:256ra135.
Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagnesis. Nat Med. 2015;21:1190–8.
Bjornsson HT. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–81.
Jones WD, Dafou D, McEntagart M, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91:358–64.
Miyake N, Tsurusaki Y, Koshimizu E, et al. Delineation of clinical features in Wiedemann-Steiner syndrome caused by KMT2A mutations. Clin Genet. 2016;89:115–9.
Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–41.
Hamosh A, Sobreira N, Hoover-Fong J, et al. PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat. 2013;34:566–71.
Adam MP, Hudgins L. Kabuki syndrome: a review. Clin Genet. 2005;67:209–19.
Dentici ML, Di Pede A, Lepri FR, et al. Kabuki syndrome: clinical and molecular diagnosis in the first year of life. Arch Dis Child. 2015;100:158–64.
Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36:425–31.
The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65.
Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6.
Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.
Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9.
Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587.
Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86.
Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31.
Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–75.
Ehrlich M, Jackson K, Weemaes C. Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF). Orphanet J Rare Dis. 2006;1:2.
van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol. 2013;33:2067–77.
Mendelsohn BA, Pronold M, Long R, Smaoui N, Slavotinek AM. Advanced bone age in a girl with Wiedemann-Steiner syndrome and an exonic deletion in KMT2A (MLL). Am J Med Genet A. 2014;8:2079–83.
Stellacci E, Onesimo R, Bruselles A, et al. Congenital immunodeficiency in an individual with Wiedemann-Steiner syndrome due to a novel missense mutation in KMT2A. Am J Med Genet A. 2016;170:2389–93.
Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acid Res. 2015;43:e47.
Barfield RT, Almli LM, Kilaru V, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–41.
Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–9.
Yuan B, Pehlivan D, Karaca E, et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest. 2015;125:636–51.
Parenti I, Teresa-Rodrigo ME, Pozojevic J, et al. Mutations in chromatin regulators functionally link Cornelia de Lange syndrome and clinically overlapping phenotypes. Hum Genet. 2017;136:307–20.
Guo C, Chang CC, Wortham M, et al. Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways. Proc Natl Acad Sci USA. 2012;109:17603–8.
Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281:15714–20.
Butcher DT, Cytrynbaum C, Turinsky AL, et al. CHARGE and Kabuki Syndromes: Gene-specific DNA Methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am J Hum Genet. 2017;100:773–88.
Choufani S, Cytrynbaum C, Chung BH, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207.
Hood RL, Schenkel LC, Nikkel SM, et al. The defining DNA methylation signature of Floating-Harbor syndrome. Sci Rep. 2016;38803. https://doi.org/10.1038/srep38803.
Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–57.
Hu JL, Zhou BO, Zhang RR, et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci USA. 2009;106:22187–92.
Wang J, Hevi S, Kurash JK, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9.
Jin B, Tao Q, Peng J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709.
Acknowledgements
We would like to thank all the families that participated in this study. We also thank Maggie Baker for assistance in selecting the candidate genes for the amplicon study. This work was supported by a grant to H.T.B. by the NIH Director’s Early Independence Award (DP5OD017877) and a grant to D.V. from the National Human Genome Research Institute (1U54HG006493).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Nara Sobreira, Martha Brucato, Li Zhang, and Christine Ladd-Acosta contributed equally to this work.
Vera A Meloni and Hans T Bjornsson contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Sobreira, N., Brucato, M., Zhang, L. et al. Patients with a Kabuki syndrome phenotype demonstrate DNA methylation abnormalities. Eur J Hum Genet 25, 1335–1344 (2017). https://doi.org/10.1038/s41431-017-0023-0
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-017-0023-0
This article is cited by
-
A KDM6 A variant in a Chinese female patient with diabetes mellitus and oligomenorrhea: a case report
Journal of Medical Case Reports (2025)
-
Immunological Aspects of Kabuki Syndrome: A Retrospective Multicenter Study of the Italian Primary Immunodeficiency Network (IPINet)
Journal of Clinical Immunology (2024)
-
Peripheral blood DNA methylation and neuroanatomical responses to HDACi treatment that rescues neurological deficits in a Kabuki syndrome mouse model
Clinical Epigenetics (2023)
-
Identification of unique DNA methylation sites in Kabuki syndrome using whole genome bisulfite sequencing and targeted hybridization capture followed by enzymatic methylation sequencing
Journal of Human Genetics (2022)
-
Interplay between chromatin marks in development and disease
Nature Reviews Genetics (2022)