Abstract
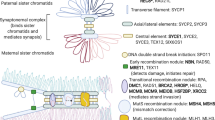
Next-generation sequencing (NGS) is increasingly being used in a clinical setting for the molecular diagnosis of patients with heterogeneous disorders, such as premature ovarian insufficiency (POI). We performed NGS of ~1000 candidate genes in four unrelated patients with POI. We discovered the genetic cause of “isolated” POI in two cases, both of which had causative variants in surprising genes. In the first case, a homozygous nonsense variant in NBN was causative. Recessive function-altering NBN variants typically cause Nijmegen breakage syndrome characterized by microcephaly, cancer predisposition, and immunodeficiency, none of which are evident in the patient. At a cellular level, we found evidence of chromosomal instability. In the second case, compound heterozygous variants in EIF2B2 were causative. Recessive EIF2B2 function-altering variants usually cause leukoencephalopathy with episodic decline. Subsequent MRI revealed subclinical neurological abnormalities. These cases demonstrate that variants in NBN and EIF2B2, which usually cause severe syndromes, can cause apparently isolated POI, and that (1) NGS can precede clinical diagnosis and guide patient management, (2) NGS can redefine the phenotypic spectrum of syndromes, and (3) NGS may make unanticipated diagnoses that must be sensitively communicated to patients. Although there is rigorous debate about the handling of secondary/incidental findings using NGS, there is little discussion of the management of causative pleiotropic gene variants that have broader implications than that for which genetic studies were sought.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Haller-Kikkatalo K, Uibo R, Kurg A, Salumets A. The prevalence and phenotypic characteristics of spontaneous premature ovarian failure: a general population registry-based study. Hum Reprod. 2015;30:1229–38.
Tucker EJ, Grover SR, Bachelot A, Touraine P, Sinclair AH. Premature ovarian insufficiency: new perspectives on genetic cause and phenotypic spectrum. Endocr Rev. 2016;37:609–35.
Eggers S, Sadedin S, van den Bergen JA, et al. Disorders of sex development: insights from targeted gene sequencing of a large international patient cohort. Genome Biol. 2016;17:243.
Varon R, Dutrannoy V, Weikert G, et al. Mild Nijmegen breakage syndrome phenotype due to alternative splicing. Hum Mol Genet. 2006;15:679–89.
Sadedin SP, Dashnow H, James PA, et al. Cpipe: a shared variant detection pipeline designed for diagnostic settings. Genome Med. 2015;7:68.
Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74.
ESP NGESP. Exome variant server. 2015. http://evs.gs.washington.edu/EVS/.
ExAC EAC. Exome aggregation consortium. 2015. http://exac.broadinstitute.org.
Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Chrzanowska KH, Szarras-Czapnik M, Gajdulewicz M, et al. High prevalence of primary ovarian insufficiency in girls and young women with Nijmegen breakage syndrome: evidence from a longitudinal study. J Clin Endocrinol Metab. 2010;95:3133–40.
Steffen J, Varon R, Mosor M, et al. Increased cancer risk of heterozygotes with NBS1 germline mutations in Poland. Int J Cancer. 2004;111:67–71.
Varon R, Reis A, Henze G, von Einsiedel HG, Sperling K, Seeger K. Mutations in the Nijmegen Breakage Syndrome gene (NBS1) in childhood acute lymphoblastic leukemia (ALL). Cancer Res. 2001;61:3570–2.
Varon R, Vissinga C, Platzer M, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–76.
Carney JP, Maser RS, Olivares H, et al. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–86.
Ranganathan V, Heine WF, Ciccone DN, et al. Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Curr Biol. 2001;11:962–6.
Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210.
Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–91.
Kobayashi J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res. 2004;45:473–8.
Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–9.
Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–21.
Ormerod E, Belengeanu V, Stoian M, et al. Nijmegen breakage syndrome: clinico-cytogenetic pattern. J Pediatr. 2009;7:19–24.
Zhang H, Dai L, Chen N, et al. Fifteen novel EIF2B1-5 mutations identified in Chinese children with leukoencephalopathy with vanishing white matter and a long term follow-up. PLoS ONE. 2015;10:e0118001.
Leegwater PA, Vermeulen G, Konst AA, et al. Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat Genet. 2001;29:383–8.
Fogli A, Schiffmann R, Bertini E, et al. The effect of genotype on the natural history of eIF2B-related leukodystrophies. Neurology. 2004;62:1509–17.
Pavitt GD. eIF2B, a mediator of general and gene-specific translational control. Biochem Soc Trans. 2005;33:1487–92.
Rowlands AG, Panniers R, Henshaw EC. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988;263:5526–33.
van der Knaap MS, Leegwater PA, Konst AA, et al. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann Neurol. 2002;51:264–70.
Pronk JC, van Kollenburg B, Scheper GC, van der Knaap MS. Vanishing white matter disease: a review with focus on its genetics. Ment Retard Dev Disabil Res Rev. 2006;12:123–8.
Fogli A, Boespflug-Tanguy O. The large spectrum of eIF2B-related diseases. Biochem Soc Trans. 2006;34:22–9.
Fogli A, Rodriguez D, Eymard-Pierre E, et al. Ovarian failure related to eukaryotic initiation factor 2B mutations. Am J Hum Genet. 2003;72:1544–50.
Warcoin M, Lespinasse J, Despouy G, et al. Fertility defects revealing germline biallelic nonsense NBN mutations. Hum Mutat. 2009;30:424–30.
Maraschio P, Peretti D, Lambiase S, et al. A new chromosome instability disorder. Clin Genet. 1986;30:353–65.
Ghezzi L, Scarpini E, Rango M, et al. A 66-year-old patient with vanishing white matter disease due to the p.Ala87Val EIF2B3 mutation. Neurology. 2012;79:2077–8.
Matsukawa T, Wang X, Liu R, et al. Adult-onset leukoencephalopathies with vanishing white matter with novel missense mutations in EIF2B2, EIF2B3, and EIF2B5. Neurogenetics. 2011;12:259–61.
Fogli A, Wong K, Eymard-Pierre E, et al. Cree leukoencephalopathy and CACH/VWM disease are allelic at the EIF2B5 locus. Ann Neurol. 2002;52:506–10.
Liu R, van der Lei HD, Wang X, et al. Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum Mutat. 2011;32:1036–45.
Labauge P, Horzinski L, Ayrignac X. et al. Natural history of adult-onset eIF2B-related disorders: a multi-centric survey of 16 cases. Brain. 2009;132:2161–9.
Fogli A, Gauthier-Barichard F, Schiffmann R, et al. Screening for known mutations in EIF2B genes in a large panel of patients with premature ovarian failure. BMC Women’s Health. 2004;4:8.
Kocarnik JM, Fullerton SM. Returning pleiotropic results from genetic testing to patients and research participants. JAMA. 2014;311:795–6.
Acknowledgements
This work was supported by a Peter Doherty Early Career Fellowship (1054432 to EJT), a National Health and Medical Research Council program grant (1074258 to AHS), a fellowship (1062854 to AHS) from the Australian National Health and Medical Research Council, and the Victorian Government’s Operational Infrastructure Support Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Tucker, E.J., Grover, S.R., Robevska, G. et al. Identification of variants in pleiotropic genes causing “isolated” premature ovarian insufficiency: implications for medical practice. Eur J Hum Genet 26, 1319–1328 (2018). https://doi.org/10.1038/s41431-018-0140-4
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0140-4
This article is cited by
-
Adult-onset vanishing white matter disease due to a novel compound heterozygous EIF2B2 mutation: a case report and brief review
Neurological Sciences (2025)
-
Next-generation sequencing of 500 POI patients identified novel responsible monogenic and oligogenic variants
Journal of Ovarian Research (2023)
-
DNA double-strand break genetic variants in patients with premature ovarian insufficiency
Journal of Ovarian Research (2023)
-
LARS2 variants can present as premature ovarian insufficiency in the absence of overt hearing loss
European Journal of Human Genetics (2023)
-
Whole-exome sequencing reveals new potential genes and variants in patients with premature ovarian insufficiency
Journal of Assisted Reproduction and Genetics (2022)