Abstract
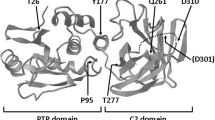
The PTEN gene encodes a master regulator protein that exerts essential functions both in the cytoplasm and in the nucleus. PTEN is mutated in the germline of both patients with heterogeneous tumor syndromic diseases, categorized as PTEN hamartoma tumor syndrome (PHTS), and a group affected with autism spectrum disorders (ASD). Previous studies have unveiled the functional heterogeneity of PTEN variants found in both patient cohorts, making functional studies necessary to provide mechanistic insights related to their pathogenicity. Here, we have functionally characterized a PTEN missense variant [c.49C>G; p.(Gln17Glu); Q17E] associated to both PHTS and ASD patients. The PTEN Q17E variant displayed partially reduced PIP3-catalytic activity and normal stability in cells, as shown using S. cerevisiae and mammalian cell experimental models. Remarkably, PTEN Q17E accumulated in the nucleus, in a process involving the PTEN N-terminal nuclear localization sequence. The analysis of additional germline-associated PTEN N-terminal variants illustrated the existence of a PTEN N-terminal region whose targeting in disease causes PTEN nuclear accumulation, in parallel with defects in PIP3-catalytic activity in cells. Our findings highlight the frequent occurrence of PTEN gene mutations targeting PTEN N-terminus whose pathogenicity may be related, at least in part, with the retention of PTEN in the nucleus. This could be important for the implementation of precision therapies for patients with alterations in the PTEN pathway.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Pulido R. PTEN: a yin-yang master regulator protein in health and disease. Methods. 2015;77-78:3–10.
Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–96.
Worby CA, Dixon JE. Pten. Annu Rev Biochem. 2014;83:641–69.
Leslie NR, Maccario H, Spinelli L, Davidson L. The significance of PTEN’s protein phosphatase activity. Adv Enzym Regul. 2009;49:190–6.
Papa AP. Protein Tyrosine Phosphatases in Cancer, pp 247–60 (Springer, New York, 2016).
Bassi C, Stambolic V. PTEN, here, there, everywhere. Cell Death Differ. 2013;20:1595–6.
Bononi A, Pinton P. Study of PTEN subcellular localization. Methods. 2015;77-78:92–103.
Gil A, Andrés-Pons A, Pulido R. Nuclear PTEN: a tale of many tails. Cell Death Differ. 2007;14:395–9.
Kreis P, Leondaritis G, Lieberam I, Eickholt BJ. Subcellular targeting and dynamic regulation of PTEN: implications for neuronal cells and neurological disorders. Front Mol Neurosci. 2014;7:23.
Gil A, Andrés-Pons A, Fernández E, et al. Nuclear localization of PTEN by a Ran-dependent mechanism enhances apoptosis: Involvement of an N-terminal nuclear localization domain and multiple nuclear exclusion motifs. Mol Biol Cell. 2006;17:4002–13.
Gil A, López JI, Pulido R. Assessing PTEN subcellular localization. Methods Mol Biol. 2016;1388:169–86.
Gil A, Rodríguez-Escudero I, Stumpf M, Molina M, Cid VJ, Pulido R. A functional dissection of PTEN N-terminus: implications in PTEN subcellular targeting and tumor suppressor activity. PLoS ONE. 2015;10:e0119287.
Andrés-Pons A, Gil A, Oliver MD, Sotelo NS, Pulido R. Cytoplasmic p27Kip1 counteracts the pro-apoptotic function of the open conformation of PTEN by retention and destabilization of PTEN outside of the nucleus. Cell Signal. 2012;24:577–87.
Odriozola L, Singh G, Hoang T, Chan AM. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem. 2007;282:23306–15.
Rahdar M, Inoue T, Meyer T, Zhang J, Vazquez F, Devreotes PN. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc Natl Acad Sci USA. 2009;106:480–5.
Vazquez F, Devreotes P. Regulation of PTEN function as a PIP3 gatekeeper through membrane interaction. Cell Cycle. 2006;5:1523–7.
Mester JE, C. PTEN hamartoma tumor syndrome. Handb Clin Neurol. 2015;132:129–37.
Kelleher RJ 3rd, Geigenmuller U, Hovhannisyan H, et al. High-throughput sequencing of mGluR signaling pathway genes reveals enrichment of rare variants in autism. PLoS ONE. 2012;7:e35003.
Orloff MS, He X, Peterson C, et al. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am J Hum Genet. 2013;92:76–80.
Riviere JB, Mirzaa GM, O’Roak BJ, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44:934–40.
Tilot AK, Frazier TW 2nd, Eng C. Balancing proliferation and connectivity in PTEN-associated autism spectrum disorder. Neurotherapeutics. 2015;12:609–19.
Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22:873–9.
Takei N, Nawa H. mTOR signaling and its roles in normal and abnormal brain development. Front Mol Neurosci. 2014;7:28.
Wei H, Alberts I, Li X. The apoptotic perspective of autism. Int J Dev Neurosci. 2014;36:13–18.
Johnston SB, Raines RT. Conformational stability and catalytic activity of PTEN variants linked to cancers and autism spectrum disorders. Biochemistry. 2015;54:1576–82.
Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, Pulido R. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20:4132–42.
Spinelli L, Black FM, Berg JN, Eickholt BJ, Leslie NR. Functionally distinct groups of inherited PTEN mutations in autism and tumour syndromes. J Med Genet. 2015;52:128–34.
Andrés-Pons A, Rodríguez-Escudero I, Gil A, et al. In vivo functional analysis of the counterbalance of hyperactive phosphatidylinositol 3-kinase p110 catalytic oncoproteins by the tumor suppressor PTEN. Cancer Res. 2007;67:9731–9.
Rodríguez-Escudero I, Fernández-Acero T, Bravo I, et al. Yeast-based methods to assess PTEN phosphoinositide phosphatase activity in vivo. Methods. 2015;77-78:172–9.
Mingo J, Erramuzpe A, Luna S, et al. One-tube-only standardized site-directed mutagenesis: an alternative approach to generate amino acid substitution collections. PLoS ONE. 2016;11:e0160972.
den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12.
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63.
Andrés-Pons A, Valiente M, Torres J, et al. Functional definition of relevant epitopes on the tumor suppressor PTEN protein. Cancer Lett. 2005;223:303–12.
Nizialek EA, Mester JL, Dhiman VK, Smiraglia DJ, Eng C. KLLN epigenotype-phenotype associations in Cowden syndrome. Eur J Human Genet. 2015;23:1538–43.
Vanderver A, Tonduti D, Kahn I, et al. Characteristic brain magnetic resonance imaging pattern in patients with macrocephaly and PTEN mutations. Am J Med Genet A. 2014;164A:627–33.
Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–79.
Denning G, Jean-Joseph B, Prince C, Durden DL, Vogt PK. A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene. 2007;26:3930–40.
Das S, Dixon JE, Cho W. Membrane-binding and activation mechanism of PTEN. Proc Natl Acad Sci USA. 2003;100:7491–6.
Lee JO, Yang H, Georgescu MM, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–34.
Nguyen HN, Yang JM, Afkari Y, et al. Engineering ePTEN, an enhanced PTEN with increased tumor suppressor activities. Proc Natl Acad Sci USA. 2014;111:E2684–2693.
Lumb CN, Sansom MS. Defining the membrane-associated state of the PTEN tumor suppressor protein. Biophys J. 2013;104:613–21.
Shenoy S, Shekhar P, Heinrich F, et al. Membrane association of the PTEN tumor suppressor: molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS ONE. 2012;7:e32591.
Bakarakos P, Theohari I, Nomikos A, et al. Immunohistochemical study of PTEN and phosphorylated mTOR proteins in familial and sporadic invasive breast carcinomas. Histopathology. 2010;56:876–82.
Yu W, He X, Ni Y, Ngeow J, Eng C. Cowden syndrome-associated germline SDHD variants alter PTEN nuclear translocation through SRC-induced PTEN oxidation. Hum Mol Genet. 2015;24:142–53.
He X, Ni Y, Wang Y, Romigh T, Eng C. Naturally occurring germline and tumor-associated mutations within the ATP-binding motifs of PTEN lead to oxidative damage of DNA associated with decreased nuclear p53. Hum Mol Genet. 2011;20:80–89.
Lobo GP, Waite KA, Planchon SM, Romigh T, Nassif NT, Eng C. Germline and somatic cancer-associated mutations in the ATP-binding motifs of PTEN influence its subcellular localization and tumor suppressive function. Hum Mol Genet. 2009;18:2851–62.
Acknowledgements
This work was supported in part by grants SAF2013-48812-R, SAF2016-79847-R, BIO2013-44112-P, and BIO2016-75030-P from Ministerio de Economía y Competitividad (Spain), and grant 2013111011 from Gobierno Vasco, Departamento de Salud (Basque Country, Spain). JM was supported by a predoctoral grant from Gobierno Vasco (Programa de Formación de Personal Investigador no doctor, Departamento de Educación, Política Lingüística y Cultura del Gobierno Vasco). SL has been the recipient of a fellowship from Bilbao Bizkaia Kutxa (BBK) (Spain). We thank Prof. Johan T. den Dunnen for providing database submission support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Mingo, J., Rodríguez-Escudero, I., Luna, S. et al. A pathogenic role for germline PTEN variants which accumulate into the nucleus. Eur J Hum Genet 26, 1180–1187 (2018). https://doi.org/10.1038/s41431-018-0155-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0155-x
This article is cited by
-
Functional analysis of PTEN variants of unknown significance from PHTS patients unveils complex patterns of PTEN biological activity in disease
European Journal of Human Genetics (2023)
-
Modeling human disease in yeast: recreating the PI3K-PTEN-Akt signaling pathway in Saccharomyces cerevisiae
International Microbiology (2020)
-
Precise definition of PTEN C-terminal epitopes and its implications in clinical oncology
npj Precision Oncology (2019)