Abstract
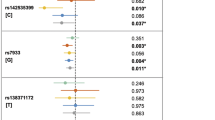
Orofacial development is a multifaceted process involving tightly regulated genetic signaling networks, that when perturbed, lead to orofacial abnormalities including cleft lip and/or cleft palate. We and others have shown an association between the cysteine-rich secretory protein LCCL domain containing 2 (CRISPLD2) gene and nonsyndromic cleft lip with or without cleft palate (NSCLP). Further, we demonstrated that knockdown of Crispld2 in zebrafish alters neural crest cell migration patterns resulting in abnormal jaw and palate development. In this study, we performed RNA profiling in zebrafish embryos and identified 249 differentially expressed genes following knockdown of Crispld2. In silico pathway analysis identified a network of seven genes previously implicated in orofacial development for which differential expression was validated in three of the seven genes (CASP8, FOS, and MMP2). Single nucleotide variant (SNV) genotyping of these three genes revealed significant associations between NSCLP and FOS/rs1046117 (GRCh38 chr14:g.75746690 T > C, p = 0.0005) in our nonHispanic white (NHW) families and MMP2/rs243836 (GRCh38 chr16:g.55534236 G > A; p = 0.002) in our Hispanic families. Nominal association was found between NSCLP and CASP8/rs3769825 (GRCh38 chr2:g.202111380 C > A; p < 0.007). Overtransmission of MMP2 haplotypes were identified in the Hispanic families (p < 0.002). Significant gene–gene interactions were identified for FOS-MMP2 in the NHW families and for CASP8-FOS in the NHW simplex family subgroup (p < 0.004). Additional in silico analysis revealed a novel gene regulatory network including five of these newly identified and 23 previously reported NSCLP genes. Our results demonstrate that animal models of orofacial clefting can be powerful tools to identify novel candidate genes and gene regulatory networks underlying NSCLP.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Gorlin RJ, Cohen MM, Hennekam RCM. Syndromes of the head and neck. Fourth Edition. New York: Oxford University Press; 2001.
Johnston MC, Hassell JR, Brown KS. The embryology of cleft lip and cleft palate. Clin Plast Surg. 1975;2:195–203.
Trasler DG, Fraser FC. Handbook of teratology. New York: Plenum Press; 1977.
Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:spc1.
Nanci A. Ten cate’s oral histology—development, structure, and function. 6th edn. Los Angelos: Mosby; 2003.
Senders CW, Peterson EC, Hendrickx AG, Cukierski MA. Development of the upper lip. Arch Facial Plast Surg. 2003;5:16–25.
Parker SE, Mai CT, Canfield MA, et al. Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 2010;88:1008–16.
Hashmi SS, Waller DK, Langlois P, Canfield M, Hecht JT. Prevalence of nonsyndromic oral clefts in Texas: 1995-1999. Am J Med Genet A. 2005;134:368–72.
Artama M, Auvinen A, Raudaskoski T, Isojarvi I, Isojarvi J. Antiepileptic drug use of women with epilepsy and congenital malformations in offspring. Neurology. 2005;64:1874–8.
Azarbayjani F, Danielsson BR. Phenytoin-induced cleft palate: evidence for embryonic cardiac bradyarrhythmia due to inhibition of delayed rectifier K+ channels resulting in hypoxia-reoxygenation damage. Teratology. 2001;63:152–60.
Lidral AC, Moreno LM. Progress toward discerning the genetics of cleft lip. Curr Opin Pediatr. 2005;17:731–9.
Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–78.
Beaty TH, Marazita ML, Leslie EJ. Genetic factors influencing risk to orofacial clefts: today’s challenges and tomorrow’s opportunities. F1000Res. 2016;5:2800.
Khandelwal KD, van Bokhoven H, Roscioli T, Carels CE, Zhou H. Genomic approaches for studying craniofacial disorders. Am J Med Genet C Semin Med Genet. 2013;163C:218–31.
Ludwig KU, Mangold E, Herms S, et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44:968–71.
Ferretti E, Li B, Zewdu R, et al. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell. 2011;21:627–41.
Liu H, Leslie EJ, Jia Z, et al. Irf6 directly regulates Klf17 in zebrafish periderm and Klf4 in murine oral epithelium, and dominant-negative KLF4 variants are present in patients with cleft lip and palate. Hum Mol Genet. 2015;25:766–76.
Blanton SH, Cortez A, Stal S, Mulliken JB, Finnell RH, Hecht JT. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137:259–62.
Ghassibe M, Bayet B, Revencu N, et al. Interferon regulatory factor-6: a gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur J Hum Genet. 2005;13:1239–42.
Scapoli L, Palmieri A, Martinelli M, et al. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–3.
Srichomthong C, Siriwan P, Shotelersuk V. Significant association between IRF6 820G- A and non-syndromic cleft lip with or without cleft palate in the Thai population. J Med Genet. 2005;42:e46.
Zucchero TM, Cooper ME, Maher BS, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–80.
Chiquet BT, Blanton SH, Burt A, et al. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet. 2008;17:2212–8.
Leslie EJ, Murray JC. Evaluating rare coding variants as contributing causes to non-syndromic cleft lip and palate. Clin Genet. 2013;84:496–500.
Letra A, Menezes R, Cooper ME, et al. CRISPLD2 variants including a C471T silent mutation may contribute to nonsyndromic cleft lip with or without cleft palate. Cleft Palate-Craniofacial J. 2011;48:363–70.
Shen X, Liu RM, Yang L, et al. The CRISPLD2 gene is involved in cleft lip and/or cleft palate in a Chinese population. Birth Defects Res A Clin Mol Teratol. 2011;91:918–24.
Shi J, Jiao X, Song T, Zhang B, Qin C, Cao F. CRISPLD2 polymorphisms are associated with non-syndromic cleft lip with or without cleft palate in a northern Chinese population. Eur J Oral Sci. 2010;118:430–3.
Chiquet BT, Lidral AC, Stal S, et al. CRISPLD2: a novel NSCLP candidate gene. Hum Mol Genet. 2007;16:2241–8.
Gibbs GM, Roelants K, O’Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocr Rev. 2008;29:865–97.
Zhang H, Sweezey NB, Kaplan F. LGL1 modulates proliferation, apoptosis, and migration of human fetal lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2015;308:L391–402.
Yuan Q, Chiquet BT, Devault L, et al. Craniofacial abnormalities result from knock down of nonsyndromic clefting gene, crispld2, in zebrafish. Genesis. 2012;50:871–81.
Swindell EC, Yuan Q, Maili LE, Tandon B, Wagner DS, Hecht JT. Crispld2 is required for neural crest cell migration and cell viability during zebrafish craniofacial development. Genesis. 2015;53:660–7.
Westerfield M: The zebrafish book: a guide for the laboratory use of zebrafish (danio rerio). Eugene: University of Oregon Press; 1995.
Calvano SE, Xiao W, Richards DR, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–7.
Becker KG, Hosack DA, Dennis G Jr, et al. PubMatrix: a tool for multiplex literature mining. BMC Bioinforma. 2003;4:61.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
Chiquet BT, Hashmi SS, Henry R, et al. Genomic screening identifies novel linkages and provides further evidence for a role of MYH9 in nonsyndromic cleft lip and palate. Eur J Human Genet. 2009;17:195–204.
Chiquet BT, Henry R, Burt A, et al. Nonsyndromic cleft lip and palate: CRISPLD genes and the folate gene pathway connection. Birth Defects Res A Clin Mol Teratol. 2011;91:44–9.
O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66.
Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42.
Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–94.
Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Human Hered. 2006;61:189–99.
Reichmann E, Schwarz H, Deiner EM, et al. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 1992;71:1103–16.
Berndt JD, Clay MR, Langenberg T, Halloran MC. Rho-kinase and myosin II affect dynamic neural crest cell behaviors during epithelial to mesenchymal transition in vivo. Dev Biol. 2008;324:236–44.
Kang P, Svoboda KK. Epithelial-mesenchymal transformation during craniofacial development. J Dent Res. 2005;84:678–90.
Blavier L, Lazaryev A, Groffen J, Heisterkamp N, DeClerck YA, Kaartinen V. TGF-beta3-induced palatogenesis requires matrix metalloproteinases. Mol Biol Cell. 2001;12:1457–66.
Brown NL, Yarram SJ, Mansell JP, Sandy JR. Matrix metalloproteinases have a role in palatogenesis. J Dent Res. 2002;81:826–30.
Smane L, Pilmane M, Akota I. Apoptosis and MMP-2, TIMP-2 expression in cleft lip and palate. Stomatologija. 2013;15:129–34.
Blanton SH, Bertin T, Serna ME, Stal S, Mulliken JB, Hecht JT. Association of chromosomal regions 3p21.2, 10p13, and 16p13.3 with nonsyndromic cleft lip and palate. Am J Med Genet A. 2004;125:23–27.
Letra A, da Silva RA, Menezes R, et al. Studies with MMP9 gene promoter polymorphism and nonsyndromic cleft lip and palate. Am J Med Genet A. 2007;143:89–91.
Letra A, Silva RA, Menezes R, et al. MMP gene polymorphisms as contributors for cleft lip/palate: association with MMP3 but not MMP1. Arch Oral Biol. 2007;52:954–60.
Letra A, Zhao M, Silva RM, Vieira AR, Hecht JT. Functional significance of MMP3 and TIMP2 polymorphisms in cleft lip/palate. J Dent Res. 2014;93:651–6.
Nikopensius T, Kempa I, Ambrozaityte L, et al. Variation in FGF1, FOXE1, and TIMP2 genes is associated with nonsyndromic cleft lip with or without cleft palate. Birth Defects Res A Clin Mol Teratol. 2011;91:218–25.
Machado RA, Messetti AC, de Aquino SN, et al. Association between genes involved in craniofacial development and nonsyndromic cleft lip and/or palate in the Brazilian population. Cleft Palate Craniofac J. 2015;53:550–6.
Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–62.
Ghavami S, Hashemi M, Ande SR, et al. Apoptosis and cancer: mutations within caspase genes. J Med Genet. 2009;46:497–510.
Dunkhase E, Ludwig KU, Knapp M, et al. Nonsyndromic cleft lip with or without cleft palate and cancer: evaluation of a possible common genetic background through the analysis of GWAS data. Genom Data. 2016;10:22–29.
Menezes R, Marazita ML, Goldstein McHenry T, et al. AXIS inhibition protein 2, orofacial clefts and a family history of cancer. J Am Dent Assoc. 2009;140:80–84.
Letra A, Menezes R, Granjeiro JM, Vieira AR. AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts. Birth Defects Res A Clin Mol Teratol. 2009;85:169–73.
Andrade Filho PA, Letra A, Cramer A, et al. Insights from studies with oral cleft genes suggest associations between WNT-pathway genes and risk of oral cancer. J Dent Res. 2011;90:740–6.
Frebourg T, Oliveira C, Hochain P, et al. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J Med Genet. 2006;43:138–42.
Zhang YJ, Zhong XP, Chen Y, Liu SR, Wu G, Liu YF. Association between CASP-8 gene polymorphisms and cancer risk in some Asian population based on a HuGE review and meta-analysis. Genet Mol Res. 2013;12:6466–76.
Cvjetkovic N, Maili L, Weymouth KS, et al. Regulatory variant in FZD6 gene contributes to nonsyndromic cleft lip and palate in an African-American family. Mol Genet Genom Med. 2015;3:440–51.
Westerman K, L. Maili, R. Plant, A. Letra J.T. Hecht, and B.T. Chiquet. “Association of β-catenin-mediated Wnt Pathway Genes with Non-syndromic Cleft Lip with or without Cleft Palate.” American Association of Dental Research. Los Angeles, CA. March 18, 2016.
Acknowledgements
We thank Maria Elena Serna and Rosa Martinez for expert clinical coordination and for managing the databases. We also thank all of the NSCLP families who kindly participated in this study. This study was approved by the Committee for the Protection of Human Subjects at UTHealth (HSC-MS-03-090) and the Animal Welfare Committee at UTHealth (AWC-14-0032). This work was funded by NIH grant R01-DE011931 to JTH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chiquet, B.T., Yuan, Q., Swindell, E.C. et al. Knockdown of Crispld2 in zebrafish identifies a novel network for nonsyndromic cleft lip with or without cleft palate candidate genes. Eur J Hum Genet 26, 1441–1450 (2018). https://doi.org/10.1038/s41431-018-0192-5
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0192-5
This article is cited by
-
The genetic factors contributing to the risk of cleft lip-cleft palate and their clinical utility
Oral and Maxillofacial Surgery (2022)
-
The functional variant of NTN1 contributes to the risk of nonsyndromic cleft lip with or without cleft palate
European Journal of Human Genetics (2020)