Abstract
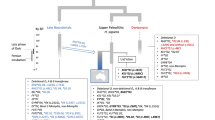
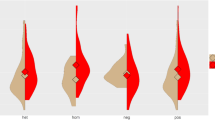
Basques show specific cultural, demographic, and genetic characteristics that have placed them as an isolated and unique population within Europe, such as their non-Indo-European language, Euskara. They have historically lived along the Western Pyrenees, between Spain and France, in one of the most important European glacial refugia during the Last Glacial Maximum. The most striking genetic characteristic is their highest frequency of the RhD blood group negative allele, a variant related to the hemolytic disease of the newborn. Both demographic and adaptive processes have been suggested as possible causes of the high frequency of RhD negative in Basques, but neither hypothesis has been clearly demonstrated. While previous studies on the Rh system in Basques have been mostly focused on serological and genotyping diversity, in this work we analyze genotyping and next generation sequencing data in order to provide a general framework of the genetic scenario of the system in Basques. In particular, we genotyped the most relevant variants of the system (D/d, E/e, and C/c), and sequenced three ~6 kb flanking regions surrounding the Rh genes in Basques and also in other populations for comparison. Our results are in agreement with previous studies, with Basques presenting the highest frequency of the RHD deletion (47.2%). Haplotype analyses of D/d, E/e, and C/c variants confirmed an association between the RhC allele, previously suggested to be under positive selection, and the RhD positive variant in non-sub-Saharan populations, including Basques. We also found extreme differentiation for the C/c variant when comparing sub-Saharan to non-sub-Saharan populations.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Aguirre A, Vicario A, Mazón LI, et al. Are the Basques a single and a unique population? Am J Hum Genet. 1991;49:450–8.
Izagirre N, de la Rúa C. An mtDNA analysis in ancient Basque populations: implications for haplogroup V as a marker for a major paleolithic expansion from southwestern europe. Am J Hum Genet. 1999;65:199–207.
Torroni A, Bandelt HJ, D’Urbano L, et al. mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet. 1998;62:1137–52.
Behar DM, Harmant C, Manry J, et al. The Basque paradigm: genetic evidence of a maternal continuity in the Franco-Cantabrian region since pre-neolithic times. Am J Hum Genet. 2012;90:486–93.
Cavalli-Sforza L. The Basque population and ancient migrations in Europe. MUNIBE (Antropol Y Arqueol. 1988;6(Suppl):129–37.
Günther T, Valdiosera C, Malmström H, et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci. 2015;112:11917–22.
Lazaridis I, Patterson N, Mittnik A, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–13.
Cavalli-Sforza L, Menozzi P, Piazza A. The history and geography of human genes. Princeton, NJ: Princeton University Press; 1994. P.
Rodríguez-Ezpeleta N, Álvarez-Busto J, Imaz L, et al. High-density SNP genotyping detects homogeneity of Spanish and French Basques, and confirms their genomic distinctiveness from other European populations. Hum Genet. 2010;128:113–7.
Laayouni H, Calafell F, Bertranpetit J. A genome-wide survey does not show the genetic distinctiveness of Basques. Hum Genet. 2010;127:455–8.
Van Der Heide HM, Magnée W, Van Loghem JJ. Blood group distribution in Basques. Am J Hum Genet. 1951;3:356–61.
Touinssi M, Chiaroni J, Degioanni A, De Micco P, Dutour O, Bauduer F. Distribution of Rhesus blood group system in the French Basques: a reappraisal using the allele-specific primers PCR method. Hum Hered. 2004;58:69–72.
Mourant AE. The distribution of the human blood groups. Oxford, UK: Blackwell Scientific Publications; 1954.
Avent ND, Reid ME. The Rh blood group system: a review. Blood. 2000;95:375–87.
Wagner FF, Flegel Wa. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–8.
Westhoff CM, Siegel DL. Rh and LW blood group antigens. In: Rossi’s principles of transfusion medicine. Chichester, WestSussex: John Wiley & Sons, Ltd.; 2016. p. 176–84.
Flegel WA The genetics of the Rhesus blood group system. In: Blood transfusion. 2007. p. 50–57.
Westhoff CM. The structure and function of the Rh antigen complex. Semin Hematol. 2007;44:42–50.
Ahmad R, de Hass M. Prevention of haemolytic disease of the fetus and newborn with reference to Anti-D. MedCrave Group LLC; 2017.
Prevention of haemolytic diseases of the newborn due to anti-D Infective endocarditis. Br Med J (Clinical Res Ed) 1981; 282:676–7.
Vogel F, Motulsky AG. Human genetics. Berlin: Springer; 1997. https://doi.org/10.1007/978-3-662-03356-2.
Etcheverry MA. El factor rhesus: Su genética y su importancia clínica. Dia Med. 1945;17:1237–59.
Perry GH, Xue Y, Smith RS, et al. Evolutionary genetics of the human Rh blood group system. Hum Genet. 2012;131:1205–16.
Kent WJ, Sugnet CW, Furey TS, et al. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–81.
O’Connell J, Gurdasani D, Delaneau O, et al. A General Approach for Haplotype Phasing across the Full Spectrum of Relatedness. PLoS Genet. 2014;10:e1004234.
Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5.
Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9.
Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48.
Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: The impact of rare variants. Genome Res. 2013;23:1514–21.
Di Gaetano C, Fiorito G, Ortu MF, et al. Sardinians genetic background explained by runs of homozygosity and genomic regions under positive selection. PLoS ONE. 2014;9:e91237 https://doi.org/10.1371/journal.pone.0091237
Gibbs RA, Belmont JW, Hardenbol P, et al. The International HapMap Project. Nature. 2003;426:789–96.
Arauna LR, Mendoza-Revilla J, Mas-Sandoval A et al. Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Mol Biol Evol. 2017;34:318–29.
Henn BM, Botigué LR, Gravel S, et al. Genomic Ancestry of North Africans Supports Back-to-Africa Migrations. PLoS Genet. 2012;8:e1002397.
Daniels G. Human blood groups. 3rd ed. 2013 https://doi.org/10.1002/9781118493595.
Carritt B, Kemp TJ, Poulter M. Evolution of the human RH (rhesus) blood group genes: A 50 year old prediction (partially) fulfilled. Hum Mol Genet. 1997;6:843–50.
Sheeladevi CS, Suchitha S, Manjunath GV, Murthy S. Hemolytic disease of the newborn due to anti-c isoimmunization: a case report. Indian J Hematol Blood Transfus. 2013;29:155–7.
Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–5.
Xue Y, Zhang X, Huang N, et al. Population differentiation as an indicator of recent positive selection in humans: an empirical evaluation. Genetics. 2009;183:1065–77.
Calvet R, Pastor JM, Fernández R, Romero JL. Blood groups of the Cantabrian population (Spain). Hum Hered. 1992;42:120–4.
Feldman MW, Nabholz M, Bodmer WF. Evolution of the Rh polymorphism: a model for the interaction of incompatibility, reproductive compensation, and heterozygote advantage. Am J Hum Genet. 1969;21:171–93.
Flegr J. Heterozygote advantage probably maintains rhesus factor blood group polymorphism: ecological regression study. PLoS ONE. 2016;11:e0147955.
Novotná M, Havliček J, Smith AP, et al. Toxoplasma and reaction time: role of toxoplasmosis in the origin, preservation and geographical distribution of Rh blood group polymorphism. Parasitology. 2008;135:1253–61. https://doi.org/10.1017/S003118200800485X
Fijarczyk A, Babik W. Detecting balancing selection in genomes: limits and prospects. Mol Ecol. 2015;24:3529–45.
Flatz G, Brinkmann H-B. A selection model for the rhesus blood-group system. J Hum Evol. 1977;6:135–40.
Gardner M, Williamson S, Casals F, et al. Extreme individual marker FST values do not imply population-specific selection in humans: the NRG1 example. Hum Genet. 2007;121:759–62.
Acknowledgements
The authors acknowledge all the volunteers participating in the study. We thank the Genomics Facility of the “Universitat Pompeu Fabra” for their technical help. We also thank Yali Xue for providing details of the sequencing primers. A.F-B was supported with a FI fellowship from the Generalitat de Catalunya (FI-DGR 2016). This work was supported by the “Agencia Estatal de Investigación” (AEI) of the “Ministerio de Economía, Industria y Competitividad” (MINEICO) and “Fondo Europeo de Desarrollo Regional” (FEDER) with project grants CGL2016-75389-P and BFU2016-77961-P, the Unidad de Excelencia María de Maeztu (MDM-2014-0370) funded by the MINECICO, and by the Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya” with the grant GRC2014SGR866.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Flores-Bello, A., Mas-Ponte, D., Rosu, M.E. et al. Sequence diversity of the Rh blood group system in Basques. Eur J Hum Genet 26, 1859–1866 (2018). https://doi.org/10.1038/s41431-018-0232-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0232-1
This article is cited by
-
Analysis of sex-specific disease patterns associated with human lifespan
GeroScience (2025)