Abstract
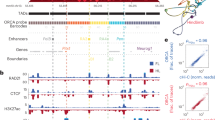
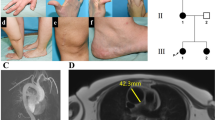
The organization of mammalian genomes into sub-megabase sized Topologically Associated Domains (TADs) has recently been revealed by techniques derived from Chromosome Conformation Capture (3 C), such as High Chromosome Contact map (Hi-C). Disruption of this organization by structural variations can lead to ectopic interactions between enhancers and promoters, and to alteration of genes expression patterns. This mechanism has already been described as the main pathophysiological mechanism in several syndromes with congenital malformations. We describe here the case of a fetus with a severe multiple congenital anomalies syndrome, including extensive polydactyly of the four limbs. This fetus carries a de novo deletion next to the IHH gene, encompassing a TAD boundary. Such an IHH TAD boundary deletion has already been described in the Dbf mouse model, which shows a quite similar, but less severe phenotype. We hypothesize that the deletion harbored by this fetus results in the same pathophysiological mechanisms as those of the Dbf model. The description of this case expands the spectrum of the disruption of chromatin architecture of WNT6/IHH/EPHA4/PAX3 locus, and could help to understand the mechanisms of chromatin interactions at this locus.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93.
Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80.
Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62:668–80.
Matharu N, Ahituv N. Minor loops in major folds: enhancer–promoter looping, chromatin restructuring, and their association with transcriptional regulation and disease. PLoS Genet. 2015;11:e1005640.
Krijger PHL, de Laat W. Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol. 2016;17:771–82.
Acemel RD, Maeso I, Gómez-Skarmeta JL. Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals: topologically associated domains. Wiley Interdiscip Rev: Dev Biol. 2017;6:e265.
Lupiáñez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25.
Greferath U, Canty AJ, Messenger J, Murphy M. Developmental expression of EphA4-tyrosine kinase receptor in the mouse brain and spinal cord. Gene Expr Patterns. 2002;2:267–74.
Helmbacher F, Schneider-Maunoury S, Topilko P, Tiret L, Charnay P. Targeting of the EphA4 tyrosine kinase receptor affects dorsal/ventral pathfinding of limb motor axons. Development. 2000;127:3313–24.
Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–22.
Labsquare Team, Schutz S, Gueudelot O. Cutepeaks 0.1.0: A simple Sanger trace file. 2017. https://doi.org/10.5281/zenodo.824555.
Firth HV, Richards SM, Bevan AP, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Human Genet. 2009;84:524–33.
Kaminsky EB, Kaul V, Paschall J et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities: Genet Med. 2011; 13: 777–84.
MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–992.
Rao SSP, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80.
Yuksel-Apak M, Bögershausen N, Pawlik B, et al. A large duplication involving the IHH locus mimics acrocallosal syndrome. Eur J Hum Genet. 2012;20:639–44.
Lyon MF, Quinney R, Glenister PH, Kerscher S, Guillot P, Boyd Y. Doublefoot: a new mouse mutant affecting development of limbs and head. Genet Res. 1996;68:221–31.
Babbs C, Furniss D, Morriss-Kay GM, Wilkie AOM. Polydactyly in the mouse mutant Doublefoot involves altered Gli3 processing and is caused by a large deletion in cis to Indian hedgehog. Mech Dev. 2008;125:517–26.
De Pace R, Skirzewski M, Damme M, et al. Altered distribution of ATG9A and accumulation of axonal aggregates in neurons from a mouse model of AP-4 deficiency syndrome. PLoS Genet. 2018;14:e1007363.
Lee T-Y, Chang W-C, Hsu JB-K, Chang T-H, Shien D-M. GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genom. 2012;13(Suppl 1):S3.
Ushijima T, Okazaki K, Tsushima H, Ishihara K, Doi T, Iwamoto Y. CCAAT/enhancer binding protein β regulates expression of indian Hedgehog during chondrocytes differentiation. PLoS ONE. 2014; 9: e104547.
Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61.
Wang Y, Zhang B, Zhang L et al. The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. 2017. https://doi.org/10.1101/112268.
Acknowledgements
The authors thank the patients for participating in this study, and the Centre of Biology and Pathology of Bordeaux University Hospital for its full support. This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical statement
Written informed consent was received for a pathological examination of fetal and placental tissues. The authors adhere to the Declaration of Helsinki Principles.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Trimouille, A., Tingaud-Sequeira, A., Pennamen, P. et al. Deletion in 2q35 excluding the IHH gene leads to fetal severe limb anomalies and suggests a disruption of chromatin architecture. Eur J Hum Genet 27, 384–388 (2019). https://doi.org/10.1038/s41431-018-0290-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0290-4