Abstract
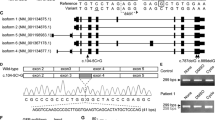
The Integrator complex subunit 1 (INTS1) is a component of the integrator complex that comprises 14 subunits and associates with RPB1 to catalyze endonucleolytic cleavage of nascent snRNAs and assist RNA polymerase II in promoter-proximal pause-release on protein-coding genes. We present five patients, including two sib pairs, with biallelic sequence variants in INTS1. The patients manifested absent or severely limited speech, an abnormal gait, hypotonia and cataracts. Exome sequencing revealed biallelic variants in INTS1 in all patients. One sib pair demonstrated a missense variant, p.(Arg77Cys), and a frameshift variant, p.(Arg1800Profs*20), another sib pair had a homozygous missense variant, p.(Pro1874Leu), and the fifth patient had a frameshift variant, p.(Leu1764Cysfs*16) and a missense variant, p.(Leu2164Pro). We also report additional clinical data on three previously described individuals with a homozygous, loss of function variant, p.(Ser1784*) in INTS1 that shared cognitive delays, cataracts and dysmorphic features with these patients. Several of the variants affected the protein C-terminus and preliminary modeling showed that the p.(Pro1874Leu) and p.(Leu2164Pro) variants may interfere with INTS1 helix folding. In view of the cataracts observed, we performed in-situ hybridization and demonstrated expression of ints1 in the zebrafish eye. We used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 to make larvae with biallelic insertion/deletion (indel) variants in ints1. The mutant larvae developed typically through gastrulation, but sections of the eye showed abnormal lens development. The distinctive phenotype associated with biallelic variants in INTS1 points to dysfunction of the integrator complex as a mechanism for intellectual disability, eye defects and craniofacial anomalies.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Baillat D, Wagner EJ. Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci. 2015;40:257–64.
Rienzo M, Casamassimi A. Integrator complex and transcription regulation: recent findings and pathophysiology. Biochim Biophys Acta. 2016;1859:1269–80.
Chen J, Waltenspiel B, Warren WD, et al. Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex. J Biol Chem. 2013;288:4867–77.
Chen J, Wagner EJ. snRNA 3’ end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38:1082–7.
Hata T, Nakayama M. Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim Biophys Acta. 2007;1773:1039–51.
Iwanami N, Okada M, Hoa V, et al. Ethylnitrosurea-induced thymus-defective mutants identify roles of KIAA1440, TRRAP, and SKIV2L2 in teleost organ development. Eur J Immunol. 2009;39:2606–16.
Oegema R, Baillat D, Schot R, et al. Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet. 2017;13:e1006809.
Depristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491498.
Mills RE, Pittard WS, Mullaney JM, et al. Natural genetic variation caused by small insertions and deletions in the human genome. Genome Res. 2011;21:830839
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucl Acids Res. 2010;38:e164.
Cingolani P. SnpEff: variant effect prediction. http://snpeff.sourceforge.net (2012).
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Schwarz JM, Rödelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6.
Teer JK, Green ED, Mullikin JC, et al. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600.
Hartley T, Wagner JD, Warman-Chardon J, et al. Whole-exome sequencing is a valuable diagnostic tool for inherited peripheral neuropathies: Outcomes from a cohort of 50 families. Clin Genet. 2018;93:301–9.
Tanaka AJ, Cho MT, Millan F, et al. Mutations in SPATA5 are Associated with Microcephaly, Intellectual Disability, Seizures, and Hearing Loss. Am J Hum Genet. 2015;97:457–64.
Zimmerman L, Stephens A, Nam SZ, et al. A completely reimplemented MPI bioinformatics toolkit with a new HHPRED server at its core. J Mol Biol. 2017;S0022-2836:30587–9.
Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815.
Chao R, Nevin L, Agarwal P, et al. A male with unilateral microphthalmia reveals a role for TMX3 in eye development. PLoS ONE. 2010;5:e10565.
Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11:583–5.
Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–7.
Al-Ghoul KJ, Lane CW, Taylor VL, et al. Distribution and type of morphological damage in human nuclear age-related cataracts. Exp Eye Res. 1996;62:237–51.
Kheirallah AK, de Moor CH, Faiz A, et al. Lung function associated gene Integrator Complex subunit 12 regulates protein synthesis pathways. BMC Genom. 2017;18:248.
Nagase T, Ishikawa K, Kikuno R, et al. Prediction of the coding sequence of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:337–45.
Dominski Z, Yang XC, Purdy M, et al. Differences and similarities between Drosophila and mammalian 3’end processing of histone pre-mRNAs. RNA. 2005;11:1835–47.
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009;25:2078–9.
Acknowledgements
We are grateful to the families for their participation. This work was supported by National Eye Institute, National Institutes of Health [grant number R21EY022779–01 to A.S.]; Erasmus Medical Center [grant Mrace #104673 to G.M.]. This work was performed under the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research (CIHR), the Ontario Genomics Institute, Ontario Research Fund, Génome Québec, and Children’s Hospital of Eastern Ontario Foundation. We also wish to acknowledge the contribution of the high-throughput sequencing platform of the McGill University and Génome Québec Innovation Centre, Montréal, Canada. R.E.L. and A.M.I. would like to thank Mary Anderson, Dr. Francois Bernier and Dr. Jillian Parboosingh for clinical and technical support, as well as helpful discussions.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
R.E.S. is an employee of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. The remaining authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Krall, M., Htun, S., Schnur, R.E. et al. Biallelic sequence variants in INTS1 in patients with developmental delays, cataracts, and craniofacial anomalies. Eur J Hum Genet 27, 582–593 (2019). https://doi.org/10.1038/s41431-018-0298-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0298-9
This article is cited by
-
The enhancer module of Integrator controls cell identity and early neural fate commitment
Nature Cell Biology (2025)
-
INTS11-related neurodevelopmental disorder: a case report and literature review
Journal of Human Genetics (2024)
-
Genomic regulation of transcription and RNA processing by the multitasking Integrator complex
Nature Reviews Molecular Cell Biology (2023)
-
BRAT1 links Integrator and defective RNA processing with neurodegeneration
Nature Communications (2022)
-
Inositol hexakisphosphate is required for Integrator function
Nature Communications (2022)