Abstract
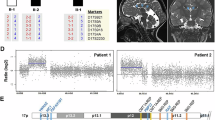
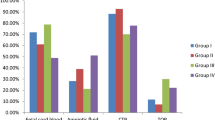
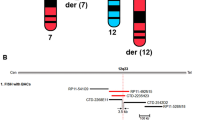
Periventricular nodular heterotopia (PNH) is a brain malformation in which nodules of neurons are ectopically retained along the lateral ventricles. Genetic causes include FLNA abnormalities (classical X-linked PNH), rare variants in ARFGEF2, DCHS1, ERMARD, FAT4, INTS8, MAP1B, MCPH1, and NEDD4L, as well as several chromosomal abnormalities. We performed array-CGH in 106 patients with different malformations of cortical development (MCD) and looked for common pathways possibly involved in PNH. Forty-two patients, including two parent/proband couples, exhibited PNH associated or not with other brain abnormalities, 44 had polymicrogyria and 20 had rarer MCDs. We found an enrichment of either large rearrangements or cryptic copy number variants (CNVs) in PNH (15/42, 35.7%) vs polymicrogyria (4/44, 9.1%) (i.e., 5.6 times increased risk for PNH of carrying a pathogenic CNV). CNVs in seven genomic regions (2p11.2q12.1, 4p15, 14q11.2q12, 16p13.3, 19q13.33, 20q13.33, 22q11) represented novel, potentially causative, associations with PNH. Through in silico analysis of genes included in imbalances whose breakpoints were clearly detailed, we detected in 9/12 unrelated patients in our series and in 15/24 previously published patients, a significant (P < 0.05) overrepresentation of genes involved in vesicle-mediated transport. Rare genomic imbalances, either small CNVs or large rearrangements, are cumulatively a frequent cause of PNH. Dysregulation of specific cellular mechanisms might play a key pathogenic role in PNH but it remains to be determined whether this is exerted through single genes or the cumulative dosage effect of more genes. Array-CGH should be considered as a first-line diagnostic test in PNH, especially if sporadic and non-classical.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Parrini E, Ramazzotti A, Dobyns WB, et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin A mutations. Brain. 2006;129:1892–906.
van Kogelenberg M, Ghedia S, McGillivray G, et al. Periventricular heterotopia in common microdeletion syndromes. Mol Syndromol. 2010;1:35–41.
Fox JW, Lamperti ED, Eksioglu YZ, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–25.
Sheen VL, Ganesh VS, Topcu M, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet. 2004;36:69–76.
Oegema R, Baillat D, Schot R, et al. Human mutations in integrator complex subunits link transcriptome integrity to brain development. PLoS Genet. 2017;13:e1006809.
Trimborn M, Bell SM, Felix C, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–6.
Cappello S, Gray MJ, Badouel C, et al. Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat Genet. 2013;45:1300–8.
Broix L, Jagline H, Ivanova E, et al. Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia. Nat Genet. 2016;48:1349–58.
Conti V, Carabalona A, Pallesi-Pocachard E, et al. Periventricular heterotopia in 6q terminal deletion syndrome: role of the C6orf70 gene. Brain. 2013;136:3378–94.
Peddibhotla S, Nagamani SC, Erez A, et al. Delineation of candidate genes responsible for structural brain abnormalities in patients with terminal deletions of chromosome 6q27. Eur J Hum Genet. 2015;23:54–60.
Heinzen EL, O’Neill AC, Zhu X, et al. De novo and inherited private variants in MAP1B in periventricular nodular heterotopia. PLoS Genet. 2018;14:e1007281.
Capra V, Biancheri R, Morana G, et al. Periventricular nodular heterotopia in Smith-Magenis syndrome. Am J Med Genet A. 2014;164A:3142–7.
Cardoso C, Boys A, Parrini E, et al. Periventricular heterotopia, mental retardation, and epilepsy associated with 5q14.3-q15 deletion. Neurology. 2009;72:784–92.
Cellini E, Disciglio V, Novara F, et al. Periventricular heterotopia with white matter abnormalities associated with 6p25 deletion. Am J Med Genet A. 2012;158A:1793–7.
Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–66.
Ciccone R, Giorda R, Gregato G, et al. Reciprocal translocations: a trap for cytogenetists? Hum Genet. 2005;117:571–82.
Kloosterman WP, Guryev V, van Roosmalen M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–24.
Guerrini R, Dobyns WB. Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 2014;13:710–26.
Wieck G, Leventer RJ, Squier WM, et al. Periventricular nodular heterotopia with overlying polymicrogyria. Brain. 2005;128:2811–21.
Sheen VL. Filamin A and Big2: a shared endocytic pathway. Bioarchitecture. 2014;4:53–57.
Sheen VL, Dixon PH, Fox JW, et al. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet. 2001;10:1775–83.
Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–8.
Bando Y, Hirano T, Tagawa Y. Dysfunction of KCNK potassium channels impairs neuronal migration in the developing mouse cerebral cortex. Cereb Cortex. 2014;24:1017–29.
Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26.
Brackenbury WJ, Isom LL. Na channel beta subunits: overachievers of the ion channel family. Front Pharmacol. 2011;2:53.
Barba C, Parrini E, Coras R, et al. Co-occurring malformations of cortical development and SCN1A gene mutations. Epilepsia. 2014;55:1009–19.
Hao YH, Fountain MD Jr, Fon Tacer K, et al. USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol Cell. 2015;59:956–69.
Fogel BL, Wexler E, Wahnich A, et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21:4171–86.
Hamada N, Ito H, Nishijo T, et al. Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep. 2016;6:30805.
Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99.
Heisterkamp N, Stam K, Groffen J, de Klein A, Grosveld G. Structural organization of the bcr gene and its role in the Ph’ translocation. Nature. 1985;315:758–61.
Park AR, Oh D, Lim SH, et al. Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a substrate of PTPRT. J Cell Sci. 2012;125:4518–31.
Narayanan AS, Reyes SB, Um K, McCarty JH, Tolias KF. The Rac-GAP Bcr is a novel regulator of the Par complex that controls cell polarity. Mol Biol Cell. 2013;24:3857–68.
Tsurusaki Y, Okamoto N, Ohashi H, et al. Coffin-Siris syndrome is a SWI/SNF complex disorder. Clin Genet. 2014;85:548–54.
Lupianez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25.
Vetro A, Dehghani MR, Kraoua L, et al. Testis development in the absence of SRY: chromosomal rearrangements at SOX9 and SOX3. Eur J Hum Genet. 2015;23:1025–32.
Kurth I, Klopocki E, Stricker S, et al. Duplications of noncoding elements 5’ of SOX9 are associated with brachydactyly-anonychia. Nat Genet. 2009;41:862–3.
den Dunnen JT, Dalgleish R, Maglott DR, et al. HGVS recommendations for the description of sequence variants: 2016 Update. Hum Mutat. 2016;37:564–9.
Acknowledgements
This work was supported by funding from the EU Seventh Framework Programme (FP7) under the project DESIRE grant agreement 602531 (to RG). We are grateful to Dr. Maria Clara Bonaglia (Cytogenetics Laboratory, Scientific Institute, IRCCS Eugenio Medea, Bosisio Parini, Lecco, Italy) and Professor Johan T. Den Dunnen (Human Genetics and Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands; curator of the HGVS nomenclature web page) for their precious help in revising the nomenclature of complex rearrangements.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cellini, E., Vetro, A., Conti, V. et al. Multiple genomic copy number variants associated with periventricular nodular heterotopia indicate extreme genetic heterogeneity. Eur J Hum Genet 27, 909–918 (2019). https://doi.org/10.1038/s41431-019-0335-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0335-3
This article is cited by
-
Neuronal hyperactivity in neurons derived from individuals with gray matter heterotopia
Nature Communications (2025)
-
Heterozygous truncating variant of TAOK1 in a boy with periventricular nodular heterotopia: a case report and literature review of TAOK1-related neurodevelopmental disorders
BMC Medical Genomics (2024)
-
Compound heterozygous variants in LAMC3 in association with posterior periventricular nodular heterotopia
BMC Medical Genomics (2021)
-
International consensus recommendations on the diagnostic work-up for malformations of cortical development
Nature Reviews Neurology (2020)