Abstract
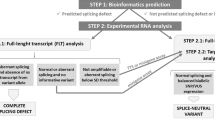
In pathogenicity assessment, RNA-based analyses are important for the correct classification of variants, and require gene-specific cut-offs for allelic representation and alternative/aberrant splicing. Beside this, the diagnostic yield of RNA-based techniques capable to detect aberrant splicing or allelic loss due to intronic/regulatory variants has to be elaborated. We established a cDNA analysis for full-length transcripts (FLT) of the four DNA mismatch repair (MMR) genes to investigate the splicing pattern and transcript integrity with active/inhibited nonsense-mediated mRNA-decay (NMD). Validation was based on results from normal controls, samples with premature termination codons (PTC), samples with splice-site defects (SSD), and samples with pathogenic putative missense variants. The method was applied to patients with variants of uncertain significance (VUS) or unexplained immunohistochemical MMR deficiency. We categorized the allelic representation into biallelic (50 ± 10%) or allelic loss (≤10%), and >10% and <40% as unclear. We defined isoforms up to 10% and exon-specific exceptions as alternative splicing, set the cut-off for SSD in cDNA + P to 30–50%, and regard >10% and <30% as unclear. FLT cDNA analyses designated 16% of all putative missense variants and 12% of VUS as SSD, detected MMR-defects in 19% of the unsolved patients, and re-classified >30% of VUS. Our method allows a standardized, systematic cDNA analysis of the MMR FLTs to assess the pathogenicity mechanism of VUS on RNA level, which will gain relevance for precision medicine and gene therapy. Diagnostic accuracy will be enhanced by detecting MMR defects in hitherto unsolved patients. The data generated will help to calibrate a high-throughput NGS-based mRNA-analysis and optimize prediction programs.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–18.
Moller P, Seppala T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2017;66:464–72.
Mangold E, Pagenstecher C, Friedl W, Mathiak M, Buettner R, Engel C, et al. Spectrum and frequencies of mutations in MSH2 and MLH1 identified in 1,721 German families suspected of hereditary nonpolyposis colorectal cancer. Int J Cancer. 2005;116:692–702.
Thompson BA, Spurdle AB, Plazzer JP, Greenblatt MS, Akagi K, Al-Mulla F, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–15.
Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–76.
Vargas-Parra GM, Gonzalez-Acosta M, Thompson BA, Gomez C, Fernandez A, Damaso E, et al. Elucidating the molecular basis of MSH2-deficient tumors by combined germline and somatic analysis. Int J Cancer. 2017;141:1365–80.
Borras E, Pineda M, Brieger A, Hinrichsen I, Gomez C, Navarro M, et al. Comprehensive functional assessment of MLH1 variants of unknown significance. Hum Mutat. 2012;33:1576–88.
Drost M, Zonneveld JB, van Hees S, Rasmussen LJ, Hofstra RM, de Wind N. A rapid and cell-free assay to test the activity of Lynch syndrome-associated MSH2 and MSH6 missense variants. Hum Mutat. 2012;33:488–94.
Hardt K, Heick SB, Betz B, Goecke T, Yazdanparast H, Kuppers R, et al. Missense variants in hMLH1 identified in patients from the German HNPCC consortium and functional studies. Fam Cancer. 2011;10:273–84.
Plotz G, Welsch C, Giron-Monzon L, Friedhoff P, Albrecht M, Piiper A, et al. Mutations in the MutSalpha interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 2006;34:6574–86.
Auclair J, Busine MP, Navarro C, Ruano E, Montmain G, Desseigne F, et al. Systematic mRNA analysis for the effect of MLH1 and MSH2 missense and silent mutations on aberrant splicing. Hum Mutat. 2006;27:145–54.
Lastella P, Surdo NC, Resta N, Guanti G, Stella A. In silico and in vivo splicing analysis of MLH1 and MSH2 missense mutations shows exon- and tissue-specific effects. BMC Genomics. 2006;7:243.
van der Klift HM, Jansen AM, van der Steenstraten N, Bik EC, Tops CM, Devilee P, et al. Splicing analysis for exonic and intronic mismatch repair gene variants associated with Lynch syndrome confirms high concordance between minigene assays and patient RNA analyses. Mol Genet Genomic Med. 2015;3:327–45.
Yamaguchi T, Wakatsuki T, Kikuchi M, Horiguchi SI, Akagi K. The silent mutation MLH1 c.543C>T resulting in aberrant splicing can cause Lynch syndrome: a case report. Jpn J Clin Oncol. 2017;47:576–80.
Baehring J, Sutter C, Kadmon M, Doeberitz MV, Gebert J. A ‘nonsense’ mutation leads to aberrant splicing of hMLH1 in a German hereditary non-polyposis colorectal cancer family. Fam Cancer. 2006;5:195–9.
Nystrom-Lahti M, Holmberg M, Fidalgo P, Salovaara R, de la Chapelle A, Jiricny J, et al. Missense and nonsense mutations in codon 659 of MLH1 cause aberrant splicing of messenger RNA in HNPCC kindreds. Genes Chromosomes Cancer. 1999;26:372–5.
Stella A, Wagner A, Shito K, Lipkin SM, Watson P, Guanti G, et al. A nonsense mutation in MLH1 causes exon skipping in three unrelated HNPCC families. Cancer Res. 2001;61:7020–4.
Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10:120–5.
Soret J, Gabut M, Tazi J. SR proteins as potential targets for therapy. Prog Mol Subcell Biol. 2006;44:65–87.
Charbonnier F, Martin C, Scotte M, Sibert L, Moreau V, Frebourg T. Alternative splicing of MLH1 messenger RNA in human normal cells. Cancer Res. 1995;55:1839–41.
Clarke LA, Jordan P, Boavida MG. Cell type specificity in alternative splicing of the human mismatch repair gene hMSH2. Eur J Hum Genet. 2000;8:347–52.
Genuardi M, Viel A, Bonora D, Capozzi E, Bellacosa A, Leonardi F, et al. Characterization of MLH1 and MSH2 alternative splicing and its relevance to molecular testing of colorectal cancer susceptibility. Hum Genet. 1998;102:15–20.
Takahashi R, Nagai K. Differences in expression between transcripts using alternative promoters of hMLH1 gene and their correlation with microsatellite instability. Oncol Rep. 2009;22:265–71.
Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM. Group IUGVW: prediction and assessment of splicing alterations: implications for clinical testing. Hum Mutat. 2008;29:1304–13.
Vreeswijk MP, van der Klift HM. Analysis and interpretation of RNA splicing alterations in genes involved in genetic disorders. Methods Mol Biol. 2012;867:49–63.
Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–9.
Thompson BA, Martins A, Spurdle AB. A review of mismatch repair gene transcripts: issues for interpretation of mRNA splicing assays. Clin Genet. 2015;87:100–8.
Davy G, Rousselin A, Goardon N, Castera L, Harter V, Legros A, et al. Detecting splicing patterns in genes involved in hereditary breast and ovarian cancer. Eur J Hum Genet. 2017;25:1147–54.
Houdayer C, Caux-Moncoutier V, Krieger S, Barrois M, Bonnet F, Bourdon V, et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012;33:1228–38.
Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–92.
Andreutti-Zaugg C, Scott RJ, Iggo R. Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res. 1997;57:3288–93.
Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8.
Morak M, Koehler U, Schackert HK, Steinke V, Royer-Pokora B, Schulmann K, et al. Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. J Med Genet. 2011;48:513–9.
Morak M, Schackert HK, Rahner N, Betz B, Ebert M, Walldorf C, et al. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008;16:804–11.
Holinski-Feder E, Muller-Koch Y, Friedl W, Moeslein G, Keller G, Plaschke J, et al. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J Biochem Biophys Methods. 2001;47:21–32.
van der Klift HM, Tops CM, Hes FJ, Devilee P, Wijnen JT. Insertion of an SVA element, a nonautonomous retrotransposon, in PMS2 intron 7 as a novel cause of Lynch syndrome. Hum Mutat. 2012;33:1051–5.
Sumitsuji I, Sugano K, Matsui T, Fukayama N, Yamaguchi K, Akasu T, et al. Frequent genomic disorganisation of MLH1 in hereditary non-polyposis colorectal cancer (HNPCC) screened by RT-PCR on puromycin treated samples. J Med Genet. 2003;40:e30.
Etzler J, Peyrl A, Zatkova A, Schildhaus HU, Ficek A, Merkelbach-Bruse S, et al. RNA-based mutation analysis identifies an unusual MSH6 splicing defect and circumvents PMS2 pseudogene interference. Hum Mutat. 2008;29:299–305.
Renkonen E, Lohi H, Jarvinen HJ, Mecklin JP, Peltomaki P. Novel splicing associations of hereditary colon cancer related DNA mismatch repair gene mutations. J Med Genet. 2004;41:e95.
Renkonen E, Zhang Y, Lohi H, Salovaara R, Abdel-Rahman WM, Nilbert M, et al. Altered expression of MLH1, MSH2, and MSH6 in predisposition to hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2003;21:3629–37.
Nomura S, Sugano K, Kashiwabara H, Taniguchi T, Fukayama N, Fujita S, et al. Enhanced detection of deleterious and other germline mutations of hMSH2 and hMLH1 in Japanese hereditary nonpolyposis colorectal cancer kindreds. Biochem Biophys Res Commun. 2000;271:120–9.
Pagenstecher C, Wehner M, Friedl W, Rahner N, Aretz S, Friedrichs N, et al. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet. 2006;119:9–22.
Tournier I, Vezain M, Martins A, Charbonnier F, Baert-Desurmont S, Olschwang S, et al. A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects. Hum Mutat. 2008;29:1412–24.
Whiley PJ, de la Hoya M, Thomassen M, Becker A, Brandao R, Pedersen IS, et al. Comparison of mRNA splicing assay protocols across multiple laboratories: recommendations for best practice in standardized clinical testing. Clin Chem. 2014;60:341–52.
Davoodi-Semiromi A, Lanyon GW, Davidson R, Connor MJ. Aberrant RNA splicing in the hMSH2 gene: molecular identification of three aberrant RNA in Scottish patients with colorectal cancer in the West of Scotland. Am J Med Genet. 2000;95:49–52.
Mori Y, Shiwaku H, Fukushige S, Wakatsuki S, Sato M, Nukiwa T, et al. Alternative splicing of hMSH2 in normal human tissues. Hum Genet. 1997;99:590–5.
Palmirotta R, Veri MC, Curia MC, Aceto G, D’Amico F, Esposito DL, et al. Transcripts with splicings of exons 15 and 16 of the hMLH1 gene in normal lymphocytes: implications in RNA-based mutation screening of hereditary non-polyposis colorectal cancer. Eur J Cancer. 1998;34:927–30.
Rhine CL, Cygan KJ, Soemedi R, Maguire S, Murray MF, Monaghan SF, et al. Hereditary cancer genes are highly susceptible to splicing mutations. PLoS Genet. 2018;14:e1007231.
Caux-Moncoutier V, Pages-Berhouet S, Michaux D, Asselain B, Castera L, De Pauw A, et al. Impact of BRCA1 and BRCA2 variants on splicing: clues from an allelic imbalance study. Eur J Hum Genet. 2009;17:1471–80.
Hesson LB, Packham D, Kwok CT, Nunez AC, Ng B, Schmidt C, et al. Lynch syndrome associated with two MLH1 promoter variants and allelic imbalance of MLH1 expression. Hum Mutat. 2015;36:622–30.
Wei Q, Bondy ML, Mao L, Gaun Y, Cheng L, Cunningham J, et al. Reduced expression of mismatch repair genes measured by multiplex reverse transcription-polymerase chain reaction in human gliomas. Cancer Res. 1997;57:1673–7.
Perera S, Bapat B. The MLH1 variants p.Arg265Cys and p.Lys618Ala affect protein stability while p.Leu749Gln affects heterodimer formation. Hum Mutat. 2008;29:332.
Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, et al. Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129:537–49.
Hinrichsen I, Brieger A, Trojan J, Zeuzem S, Nilbert M, Plotz G. Expression defect size among unclassified MLH1 variants determines pathogenicity in Lynch syndrome diagnosis. Clin Cancer Res. 2013;19:2432–41.
Koger N, Paulsen L, Lopez-Kostner F, Della Valle A, Vaccaro CA, Palmero EI, et al. Evaluation of MLH1 variants of unclear significance. Genes Chromosomes Cancer. 2018;57:350–8.
Hayward BE, De Vos M, Valleley EM, Charlton RS, Taylor GR, Sheridan E, et al. Extensive gene conversion at the PMS2 DNA mismatch repair locus. Hum Mutat. 2007;28:424–30.
Acknowledgements
We thank all patients for their participation in this study, as well as their respective doctors for contributing materials and clinical information.
Funding
This work was based on projects funded by grants from the German Cancer Aid (Deutsche Krebshilfe) (#111222) and the Wilhelm Sander-Stiftung (#2012.081.1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Morak, M., Schaefer, K., Steinke-Lange, V. et al. Full-length transcript amplification and sequencing as universal method to test mRNA integrity and biallelic expression in mismatch repair genes. Eur J Hum Genet 27, 1808–1820 (2019). https://doi.org/10.1038/s41431-019-0472-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0472-8
This article is cited by
-
Novel MSH6 exon 5–6 skipping variant in a Taiwanese family with Lynch syndrome: implications for genetic testing and cancer management
Molecular Cytogenetics (2025)
-
Whole genome sequencing completes the molecular genetic testing workflow of patients with Lynch syndrome
npj Genomic Medicine (2025)
-
A 39 kb structural variant causing Lynch Syndrome detected by optical genome mapping and nanopore sequencing
European Journal of Human Genetics (2024)
-
Splicing analyses for variants in MMR genes: best practice recommendations from the European Mismatch Repair Working Group
European Journal of Human Genetics (2022)
-
Prevalence of CNV-neutral structural genomic rearrangements in MLH1, MSH2, and PMS2 not detectable in routine NGS diagnostics
Familial Cancer (2020)