Abstract
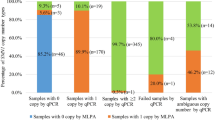
Establishing nucleic acid-based assays for genetic newborn screening (NBS) provides the possibility to screen for genetically encoded diseases like spinal muscular atrophy (SMA), best before the onset of symptoms. Such assays should be easily scalable to 384-well reactions that make the screening of up to 2000 samples per day possible. We developed a test procedure based on a cleanup protocol for dried blood spots and a quantitative (q)PCR to screen for a homozygous deletion of exon 7 of the survival of motor neuron 1 gene (SMN1) that is responsible for >95% of SMA patients. Performance of this setup is evaluated in detail and tested on routine samples. Our cleanup method for nucleic acids from dried blood spots yields enough DNA for diverse subsequent qPCR applications. To date, we have applied this approach to test 213,279 samples within 18 months. Thirty patients were identified and confirmed, implying an incidence of 1:7109 for the homozygous deletion. Using our cleanup method, a rapid workflow could be established to prepare nucleic acids from dried blood spot cards. Targeting the exon 7 deletion, no invalid, false-positive, or false-negative results were reported to date. This allows timely identification of the disease and grants access to the recently introduced treatment options, in most cases before the onset of symptoms. Carriers are not identified, thus, there are no concerns of whether to report them.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Therrell BL, Padilla CD, Loeber JG, Kneisser I, Saadallah A, Borrajo GJ, et al. Current status of newborn screening worldwide: 2015. Semin Perinatol. 2015;39:171–87.
Andermann A, Blancquaert I, Beauchamp S, Costea I. Guiding policy decisions for genetic screening: developing a systematic and transparent approach. Public Health Genom. 2011;14:9–16.
Andermann A, Blancquaert I, Beauchamp S, Dery V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ 2008;86:317–9.
Dobrow MJ, Hagens V, Chafe R, Sullivan T, Rabeneck L. Consolidated principles for screening based on a systematic review and consensus process. CMAJ 2018;190:E422–9.
Finkel RS, Mercuri E, Meyer OH, Simonds AK, Schroth MK, Graham RJ, et al. Diagnosis and management of spinal muscular atrophy: part 2: pulmonary and acute care; medications, supplements and immunizations; other organ systems; and ethics. Neuromuscul Disord. 2018;28:197–207.
Mercuri E, Finkel RS, Muntoni F, Wirth B, Montes J, Main M, et al. Diagnosis and management of spinal muscular atrophy: Part 1: Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscul Disord. 2018;28:103–15.
Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM, et al. Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet. 2012;20:27–32.
Verhaart IEC, Robertson A, Leary R, McMacken G, Konig K, Kirschner J, et al. A multi-source approach to determine SMA incidence and research ready population. J Neurol. 2017;264:1465–73.
Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–68.
Pyatt RE, Prior TW. A feasibility study for the newborn screening of spinal muscular atrophy. Genet Med. 2006;8:428–37.
Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–65.
Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat. 2000;15:228–37.
Prior TW, Snyder PJ, Rink BD, Pearl DK, Pyatt RE, Mihal DC, et al. Newborn and carrier screening for spinal muscular atrophy. Am J Med Genet A. 2010;152A:1608–16.
Wirth B, Schmidt T, Hahnen E, Rudnik-Schoneborn S, Krawczak M, Muller-Myhsok B, et al. De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet. 1997;61:1102–11.
Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schoneborn S, et al. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999;64:1340–56.
Kolb SJ, Kissel JT. Spinal muscular atrophy. Neurol Clin. 2015;33:831–46.
Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–26.
Messina S. New directions for SMA therapy. J Clin Med. 2018;7:251.
Alcantara-Ortigoza MA, Belmont-Martinez L, Vela-Amieva M, Gonzalez-Del Angel A. Analysis of the CTNS gene in nephropathic cystinosis Mexican patients: report of four novel mutations and identification of a false positive 57-kb deletion genotype with LDM-2/exon 4 multiplex PCR assay. Genet Test. 2008;12:409–14.
Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, et al. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N Engl J Med. 2018;378:625–35.
Saffari A, Kolker S, Hoffmann GF, Weiler M, Ziegler A. Novel challenges in spinal muscular atrophy - How to screen and whom to treat? Ann Clin Transl Neurol. 2019;6:197–205.
Chien YH, Chiang SC, Weng WC, Lee NC, Lin CJ, Hsieh WS, et al. Presymptomatic diagnosis of spinal muscular atrophy through newborn screening. J Pediatr. 2017;190:124–9.
Strom CM, Anderson B, Peng M, Patel U, Braastad CD, Sun W. 1000 sample comparison of MLPA and RT-PCR for carrier detection and diagnostic testing for spinal muscular atrophy type 1. Open J Genet. 2013;3:111–4.
Arkblad EL, Darin N, Berg K, Kimber E, Brandberg G, Lindberg C, et al. Multiplex ligation-dependent probe amplification improves diagnostics in spinal muscular atrophy. Neuromuscul Disord. 2006;16:830–8.
Forst HT. Problems of multiple tests and evaluations in drug research. Arzneim Forsch. 1985;35:563–9.
Phan HC, Taylor JL, Hannon H, Howell R. Newborn screening for spinal muscular atrophy: anticipating an imminent need. Semin Perinatol. 2015;39:217–29.
Durner J. Clinical chemistry: challenges for analytical chemistry and the nanosciences from medicine. Angew Chem Int Ed Engl. 2010;49:1026–51.
Saavedra-Matiz CA, Isabelle JT, Biski CK, Duva SJ, Sweeney ML, Parker AL, et al. Cost-effective and scalable DNA extraction method from dried blood spots. Clin Chem. 2013;59:1045–51.
Kraszewski JN, Kay DM, Stevens CF, Koval C, Haser B, Ortiz V, et al. Pilot study of population-based newborn screening for spinal muscular atrophy in New York state. Genet Med. 2018;20:608–13.
Taylor JL, Lee FK, Yazdanpanah GK, Staropoli JF, Liu M, Carulli JP, et al. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin Chem. 2015;61:412–9.
Ar Rochmah M, Awano H, Awaya T, Harahap NIF, Morisada N, Bouike Y, et al. Genetic screening of spinal muscular atrophy using a real-time modified COP-PCR technique with dried blood-spot DNA. Brain Dev. 2017;39:851–60.
Hao Z, Fu D, Ming Y, Yang J, Huang Q, Lin W, et al. Large scale newborn deafness genetic screening of 142,417 neonates in Wuhan, China. PLoS ONE. 2018;13:e0195740.
Smith M, Calabro V, Chong B, Gardiner N, Cowie S, du Sart D. Population screening and cascade testing for carriers of SMA. Eur J Hum Genet. 2007;15:759–66.
Calucho M, Bernal S, Alias L, March F, Vencesla A, Rodriguez-Alvarez FJ, et al. Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord. 2018;28:208–15.
Oprea GE, Krober S, McWhorter ML, Rossoll W, Muller S, Krawczak M, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–7.
Crooke ST, Witztum JL, Bennett CF, Baker BF. RNA-targeted therapeutics. Cell Metab. 2018;27:714–39.
Glascock J, Sampson J, Haidet-Phillips A, Connolly A, Darras B, Day J, et al. Treatment algorithm for infants diagnosed with spinal muscular atrophy through newborn screening. J Neuromuscul Dis. 2018;5:145–58.
Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med. 2017;377:1713–22.
Acknowledgements
We thank Peta Snikeris for proofing the paper.
Funding
The screening was funded by the German Cystinosis Foundation (Cystinose Stiftung, DSZ-Regional Office Munich, Widenmayerstr. 10, 80538 Munich, Germany). The funders had no influence on the design, interpretation, and publication of these data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Czibere, L., Burggraf, S., Fleige, T. et al. High-throughput genetic newborn screening for spinal muscular atrophy by rapid nucleic acid extraction from dried blood spots and 384-well qPCR. Eur J Hum Genet 28, 23–30 (2020). https://doi.org/10.1038/s41431-019-0476-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0476-4
This article is cited by
-
High-sensitivity qPCR detection method based on silver flower-like LSPR-active material
Scientific Reports (2025)
-
Neugeborenenscreening auf spinale Muskelatrophie
Monatsschrift Kinderheilkunde (2024)
-
Expanding the Australian Newborn Blood Spot Screening Program using genomic sequencing: do we want it and are we ready?
European Journal of Human Genetics (2023)
-
Genomisches Neugeborenenscreening – Forschungsansätze, Herausforderungen und Chancen
Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz (2023)
-
Next generation sequencing is a highly reliable method to analyze exon 7 deletion of survival motor neuron 1 (SMN1) gene
Scientific Reports (2022)