Abstract
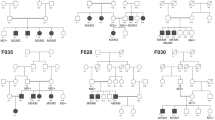
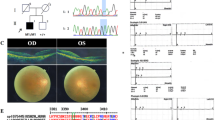
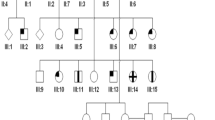
Visual impairment due to inherited ophthalmic disorders is amongst the most common disabilities observed in populations practicing consanguineous marriages. Here we investigated the molecular genetic basis of an unselected broad range of ophthalmic disorders in 20 consanguineous families from Arab villages of Israel and the Palestinian Authority. Most patients had little or very poor prior clinical workup and were recruited in a field study. Homozygosity mapping followed by candidate gene sequencing applying conventional Sanger sequencing or targeted next generation sequencing was performed in six families. In the remaining 14 families, one affected subject per family was chosen for whole exome sequencing. We discovered likely disease-causing variants, all homozygous, in 19 of 20 independent families (95%) including a previously reported novel disease gene for congenital nystagmus associated with foveal hypoplasia. Moreover, we found a family in which disease-causing variants for two collagenopathies — Stickler and Knobloch syndrome — segregate within a large sibship. Nine of the 19 distinct variants observed in this study were novel. Our study demonstrated a very high molecular diagnostic yield for a highly diverse spectrum of rare ophthalmic disorders in Arab patients from Israel and the Palestinian Authority, even with very limited prior clinical investigation. We conclude that ‘genetic testing first' may be an economic way to direct clinical care and to support proper genetic counseling and risk assessment in these families.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dale N, Salt A. Early support developmental journal for children with visual impairment: the case for a new developmental framework for early intervention. Child Care Health Dev 2007;33:684–90.
Lohmann K, Klein C. Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics. 2014;11:699–707.
Weisschuh N, Mayer AK, Strom TM, Kohl S, Glöckle N, Schubach M, et al. Mutation detection in patients with retinal dystrophies using targeted next generation sequencing. PLoS One. 2016;11:e0145951.
Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8:109–29.
Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29:335–75.
Graw J. The genetic and molecular basis of congenital eye defects. Nat Rev Genet. 2003;4:876–88.
Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper-an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–99.
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strainw1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–62.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–49.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Mayer AK, Mahajnah M, Thomas MG, Cohen Y, Habib A, Schulze M, et al. Homozygous stop mutation in AHR causes autosomal recessive foveal hypoplasia and infantile nystagmus. Brain. 2019;142:1528–34.
Kuniyoshi K, Ikeo K, Sakuramoto H, Furuno M, Yoshitake K, Hatsukawa Y, et al. Novel nonsense and splice site mutations in CRB1 gene in two Japanese patients with early-onset retinal dystrophy. Doc Ophthalmol. 2015;130:49–55.
Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;6:17.
Jaber L, Bailey-Wilson JE, Haj-Yehia M, Hernandez J, Shohat M. Consanguineous matings in an Israeli-Arab community. Arch Pediatr Adolesc Med. 1994;148:412–5.
Sharkia R, Zaid M, Athamna A, Cohen D, Azem A, Zalan A. The changing pattern of consanguinity in a selected region of the Israeli Arab community. Am J Hum Biol 2008;20:72–7.
Zobor D, Balousha G, Baumann B, Wissinger B. Homozygosity mapping reveals new nonsense mutation in the FAM161A gene causing autosomal recessive retinitis pigmentosa in a Palestinian family. Mol Vis. 2014;20:178–82.
Mayer AK, Mahajnah M, Zobor D, Bonin M, Sharkia R, Wissinger B. Novel homozygous large deletion including the 5' part of the SPATA7 gene in a consanguineous Israeli Muslim Arab family. Mol Vis. 2015;21:306–15.
Burke TR, Fishman GA, Zernant J, Schubert C, Tsang SH, Smith RT, et al. Retinal phenotypes in patients homozygous for the G1961E mutation in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012;53:4458–67.
Beryozkin A, Zelinger L, Bandah-Rozenfeld D, Harel A, Strom TA, Merin S, et al. Mutations in CRB1 are a relatively common cause of autosomal recessive early-onset retinal degeneration in the Israeli and Palestinian populations. Invest Ophthalmol Vis Sci. 2013;54:2068–75.
den Hollander AI, ten Brink JB, de Kok YJ, van Soest S, van den Born LI, van Driel MA, et al. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat Genet. 1999;23:217–21.
Lotery AJ, Jacobson SG, Fishman GA, Weleber RG, Fulton AB, Namperumalsamy P, et al. Mutations in the CRB1 gene cause Leber congenital amaurosis. Arch Ophthalmol. 2001;119:415–20.
Wissinger B, Gamer D, Jägle H, Giorda R, Marx T, Mayer S, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–37.
Besnard T, Vaché C, Baux D, Larrieu L, Abadie C, Blanchet C, et al. Non-USH2A mutations in USH2 patients. Hum Mutat. 2012;33:504–10.
Sharon D, Banin E. Nonsyndromic retinitis pigmentosa is highly prevalent in the Jerusalem region with a high frequency of founder mutations. Mol Vis. 2015;21:783–92.
Maw MA, Kennedy B, Knight A, Bridges R, Roth KE, Mani EJ, et al. Mutation of the gene encoding cellular retinaldehyde-binding protein in autosomal recessive retinitis pigmentosa. Nat Genet. 1997;17:198–200.
Morimura H, Berson EL, Dryja TP. Recessive mutations in the RLBP1 gene encoding cellular retinaldehyde-binding protein in a form of retinitis punctata albescens. Invest Ophthalmol Vis Sci. 1999;40:1000–4.
Iannaccone A, Tedesco SA, Gallaher KT, Yamamoto H, Charles S, Dryja TP. Fundus albipunctatus in a 6-year old girl due to compound heterozygous mutations in the RDH5 gene. Doc Ophthalmol. 2007;115:111–6.
Schatz P, Preising M, Lorenz B, Sander B, Larsen M, Eckstein C, et al. Lack of autofluorescence in fundus albipunctatus associated with mutations in RDH5. Retina. 2010;30:1704–13.
Pras E, Pras E, Reznik-Wolf H, Sharon D, Raivech S, Barkana Y, et al. Fundus albipunctatus: novel mutations and phenotypic description of Israeli patients. Mol Vis. 2012;18:1712–8.
Lidén M, Romert A, Tryggvason K, Persson B, Eriksson U. Biochemical defects in 11-cis-retinol dehydrogenase mutants associated with fundus albipunctatus. J Biol Chem. 2001;276:49251–7.
Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–91.
Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, et al. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet. 2001;68:1295–8.
Hameed A, Abid A, Aziz A, Ismail M, Mehdi SQ, Khaliq S. Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy. J Med Genet. 2003;40:616–9.
Aker M, Rouvinski A, Hashavia S, Ta-Shma A, Shaag A, Zenvirt S, et al. An SNX10 mutation causes malignant osteopetrosis of infancy. J Med Genet. 2012;49:221–6.
Pangrazio A, Fasth A, Sbardellati A, Orchard PJ, Kasow KA, Raza J, et al. SNX10 mutations define a subgroup of human autosomal recessive osteopetrosis with variable clinical severity. J Bone Min Res. 2013;28:1041–9.
Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu YF, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–81.
Bar-Yosef U, Abuelaish I, Harel T, Hendler N, Ofir R, Birk OS. CHX10 mutations cause non-syndromic microphthalmia/ anophthalmia in Arab and Jewish kindreds. Hum Genet. 2004;115:302–9.
Williamson KA, FitzPatrick DR. The genetic architecture of microphthalmia, anophthalmia and coloboma. Eur J Med Genet. 2014;57:369–80.
Khan K, Rudkin A, Parry DA, Burdon KP, McKibbin M, Logan CV, et al. Homozygous mutations in PXDN cause congenital cataract, corneal opacity, and developmental glaucoma. Am J Hum Genet. 2011;89:464–73.
Choi A, Lao R, Ling-Fung Tang P, Wan E, Mayer W, Bardakjian T, et al. Novel mutations in PXDN cause microphthalmia and anterior segment dysgenesis. Eur J Hum Genet. 2015;23:337–41.
Baker S, Booth C, Fillman C, Shapiro M, Blair MP, Hyland JC, et al. A loss of function mutation in the COL9A2 gene causes autosomal recessive Stickler syndrome. Am J Med Genet A. 2011;155A:1668–72.
Nixon TRW, Alexander P, Richards A, McNinch A, Bearcrift PWP, Cobben J, et al. Homozygous Type IX collagen variants (COL9A1, COL9A2, and COL9A3) causing recessive Stickler syndrome-expanding the phenotype. Am J Med Genet A. 2019;179:1498–506.
Suzuki OT, Sertié AL, Der Kaloustian VM, Kok F, Carpenter M, Murray J, et al. Molecular analysis of collagen XVIII reveals novel mutations, presence of a third isoform, and possible genetic heterogeneity in Knobloch syndrome. Am J Hum Genet. 2002;71:1320–9.
Charsar BA, Goldberg EM. Polymicrogyria and intractable epilepsy in siblings With Knobloch syndrome and homozygous mutation of COL18A1. Pediatr Neurol. 2017;76:91–2.
Zhang LS, Li HB, Zeng J, Yang Y, Ding C. Knobloch syndrome caused by homozygous frameshift mutation of the COL18A1 gene in a Chinese pedigree. Int J Ophthalmol. 2018;11:918–22.
Stambolian D, Ai Y, Sidjanin D, Nesburn K, Sathe G, Rosenberg M, et al. Cloning of the galactokinase cDNA and identification of mutations in two families with cataracts. Nat Genet. 1995;10:307–12.
Asada M, Okano Y, Imamura T, Suyama I, Hase Y, Isshiki G. Molecular characterization of galactokinase deficiency in Japanese patients. J Hum Genet. 1999;44:377–82.
Segal S, Rutman JY, Frimpter GW. Galactokinase deficiency and mental retardation. J Pediatr. 1979;95:750–2.
Potter NL, Nievergelt Y, Shriberg LD. Motor and speech disorders in classic galactosemia. JIMD Rep. 2013;11:31–41.
Vanderschueren-Lodeweyckx M, Debruyne F, Dooms L, Eggermont E, Eeckels R. Sensorineural hearing loss in sporadic congenital hypothyroidism. Arch Dis Child. 1983;58:419–22.
Virtanen M. Manifestations of congenital hypothyroidism during the 1st week of life. Eur J Pediatr. 1988;147:270–4.
Chevallier A, Mialot A, Petit JM, Fernandez-Salguero P, Barouki R, Coumoul X, et al. Oculomotor deficits in aryl hydrocarbon receptor null mouse. PLoS One. 2013;8:e53520.
Juricek L, Carcaud J, Pelhaitre A, et al. AhR-deficiency as a cause of demyelinating disease and inflammation. Sci Rep. 2017;7:9794.
Hanein S, Perrault I, Gerber S, Tanguy G, Barbet F, Ducroq D, et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype–phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23:306–17.
Genead MA, Fishman GA, Rha J, Dubis AM, Bonci DM, Dubra A, et al. Photoreceptor structure and function in patients with congenital achromatopsia. Invest Ophthalmol Vis Sci. 2011;52:7298–308.
Acknowledgements
We are grateful to the patients and family members who participated in this study. Further, we are indebted to Dr. Ibrahim Yehya, the Scientific Director of the Triangle Regional Research and Development Center.
Funding
This work was supported by grants from the German Research Foundation (SCHO 754/5-2, WI 1189/8-2, and BA 2417/2-2) to LS, BW, and PB (principle applicants), and to GB, RS, SA, and AA (co-applicants). LS is a member of the European Network for Rare Neurological Diseases (Project ID No. 739510). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mayer, A.K., Balousha, G., Sharkia, R. et al. Unraveling the genetic cause of hereditary ophthalmic disorders in Arab societies from Israel and the Palestinian Authority. Eur J Hum Genet 28, 742–753 (2020). https://doi.org/10.1038/s41431-019-0566-3
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0566-3