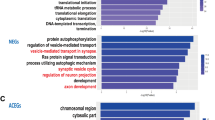
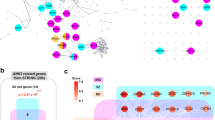
Abstract
Upon the discovery of numerous genes involved in the pathogenesis of neurodevelopmental disorders, several studies showed that a significant proportion of these genes converge on common pathways and protein networks. Here, we used a reversed approach, by screening the AnkyrinG protein-protein interaction network for genetic variation in a large cohort of 1009 cases with neurodevelopmental disorders. We identified a significant enrichment of de novo potentially disease-causing variants in this network, confirming that this protein network plays an important role in the emergence of several neurodevelopmental disorders.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
APA: American Psychiatric Association. Diagnostic and statistical manual of mental disorders DSM-V, Fifth ed., Text revision. Washington DC: American Psychiatric Association; 2013.
Oeseburg B, Dijkstra GJ, Groothoff JW, Reijneveld SA, Jansen DE. Prevalence of chronic health conditions in children with intellectual disability: a systematic literature review. Intellect Dev Dis. 2011;49:59–85.
Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, Nagamani SC, et al. Microdeletion 15q13.3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–8.
Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6.
Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75.
McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–7.
Zufferey F, Sherr EH, Beckmann ND, Hanson E, Maillard AM, Hippolyte L, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet. 2012;49:660–8.
O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science (New York, NY). 2012;338:1619–22.
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99.
de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9.
Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82.
Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94.
van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104.
Davis JQ, Lambert S, Bennett V. Molecular composition of the node of Ranvier: identification of ankyrin-binding cell adhesion molecules neurofascin (mucin+/third FNIII domain-) and NrCAM at nodal axon segments. J Cell Biol. 1996;135:1355–67.
Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–304.
Berghs S, Aggujaro D, Dirkx R Jr, Maksimova E, Stabach P, Hermel JM, et al. betaIV spectrin, a new spectrin localized at axon initial segments and nodes of ranvier in the central and peripheral nervous system. J Cell Biol. 2000;151:985–1002.
Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, et al. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–613.
Jenkins PM, Kim N, Jones SL, Tseng WC, Svitkina TM, Yin HH, et al. Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc Natl Acad Sci USA. 2015;112:957–64.
Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–9.
Sobotzik JM, Sie JM, Politi C, Del Turco D, Bennett V, Deller T, et al. AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc Natl Acad Sci USA. 2009;106:17564–9.
Smith KR, Kopeikina KJ, Fawcett-Patel JM, Leaderbrand K, Gao R, Schurmann B, et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron. 2014;84:399–415.
Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–8.
Yuan A, Yi Z, Wang Q, Sun J, Li Z, Du Y, et al. ANK3 as a risk gene for schizophrenia: New data in han Chinese and meta analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:997–1005.
Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J, et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry. 2009;14:487–91.
Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–53.
Logue MW, Solovieff N, Leussis MP, Wolf EJ, Melista E, Baldwin C, et al. The ankyrin-3 gene is associated with posttraumatic stress disorder and externalizing comorbidity. Psychoneuroendocrinology. 2013;38:2249–57.
Bi C, Wu J, Jiang T, Liu Q, Cai W, Yu P, et al. Mutations of ANK3 identified by exome sequencing are associated with autism ausceptibility. Hum Mutat. 2012;33:1635–8.
Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA, et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet. 2013;22:1960–70.
Brechet A, Fache MP, Brachet A, Ferracci G, Baude A, Irondelle M, et al. Protein kinase CK2 contributes to the organization of sodium channels in axonal membranes by regulating their interactions with ankyrin G. J Cell Biol. 2008;183:1101–14.
Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50.
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13.
Okur V, Cho MT, Henderson L, Retterer K, Schneider M, Sattler S, et al. De novo mutations in CSNK2A1 are associated with neurodevelopmental abnormalities and dysmorphic features. Hum Genet. 2016;135:699–705.
Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Sarno S, Vaglio P, Meggio F, Issinger OG, Pinna LA. Protein kinase CK2 mutants defective in substrate recognition. Purification and kinetic analysis. J Biol Chem. 1996;271:10595–601.
Chiu ATG, Pei SLC, Mak CCY, Leung GKC, Yu MHC, Lee SL, et al. Okur-Chung neurodevelopmental syndrome: Eight additional cases with implications on phenotype and genotype expansion. Clin Genet. 2018;93:880–90.
Akahira-Azuma M, Tsurusaki Y, Enomoto Y, Mitsui J, Kurosawa K. Refining the clinical phenotype of Okur-Chung neurodevelopmental syndrome. Hum Genome Var. 2018;5:18011.
Wallace RH, Wang DW, Singh R, Scheffer IE, George AL Jr, Phillips HA, et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19:366–70.
Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6.
Patino GA, Isom LL. Electrophysiology and beyond: multiple roles of Na+ channel beta subunits in development and disease. Neurosci Lett. 2010;486:53–9.
Scheffer IE, Harkin LA, Grinton BE, Dibbens LM, Turner SJ, Zielinski MA, et al. Temporal lobe epilepsy and GEFS+ phenotypes associated with SCN1B mutations. Brain: J Neurol. 2007;130:100–9.
McCormick KA, Isom LL, Ragsdale D, Smith D, Scheuer T, Catterall WA. Molecular determinants of Na+ channel function in the extracellular domain of the beta1 subunit. J Biol Chem. 1998;273:3954–62.
Malhotra JD, Koopmann MC, Kazen-Gillespie KA, Fettman N, Hortsch M, Isom LL. Structural requirements for interaction of sodium channel beta 1 subunits with ankyrin. J Biol Chem. 2002;277:26681–8.
Fry AE, Rees E, Thompson R, Mantripragada K, Blake P, Jones G, et al. Pathogenic copy number variants and SCN1A mutations in patients with intellectual disability and childhood-onset epilepsy. BMC Med Genet. 2016;17:34.
Weckhuysen S, Mandelstam S, Suls A, Audenaert D, Deconinck T, Claes LRF, et al. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol. 2012;71:15–25.
Papadimitriou S, Gazzo A, Versbraegen N, Nachtegael C, Aerts J, Moreau Y, et al. Predicting disease-causing variant combinations. Proc Natl Acad Sci USA. 2019;116:11878–87.
Vissers LELM, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12.
Beyens M, Boeckx N, Van Camp G, Op de Beeck K, Vandeweyer G. pyAmpli: an amplicon-based variant filter pipeline for targeted resequencing data. BMC Bioinform. 2017;18:554.
Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18.
Vandeweyer G, Helsmoortel C, Van Dijck A, Vulto-van Silfhout AT, Coe BP, Bernier R. et al. The Transcriptional Regulator ADNP Links the BAF (SWI/SNF) Complexes With Autism. Am J Med Genet C. 2014;166:315–26.
Kosho T, Okamoto N, Ohashi H, Tsurusaki Y, Imai Y, Hibi-Ko Y, et al. Clinical correlations of mutations affecting six components of the SWI/SNF complex: detailed description of 21 patients and a review of the literature. Am J Med Genet. 2013;161A:1221–37.
Braat S, Kooy RF. Fragile X syndrome neurobiology translates into rational therapy. Drug Discov Today. 2014;19:510–9.
Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB Jr, Moine H, Kooy RF, et al. Fragile X syndrome. Nat Rev Dis Prim. 2017;3:17065.
Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15.
Boyle EA, O’Roak BJ, Martin BK, Kumar A, Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30:2670–2.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4.
Acknowledgements
This work was sponsored by a grant from the Fonds Wetenschappelijk Onderzoek - Vlaanderen (FWO) to RFK and GVDW and by a grant from the Netherlands Organization for Health Research and Development (912-12-109) to LELMV and BBAdV.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
van der Werf, I.M., Jansen, S., de Vries, P.F. et al. Overrepresentation of genetic variation in the AnkyrinG interactome is related to a range of neurodevelopmental disorders. Eur J Hum Genet 28, 1726–1733 (2020). https://doi.org/10.1038/s41431-020-0682-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-020-0682-0
This article is cited by
-
Deciphering the molecular landscape of NMDAR-E associated ovarian teratomas: a Multi-Omics approach
Journal of Ovarian Research (2025)