Abstract
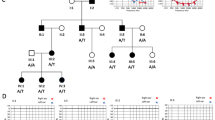
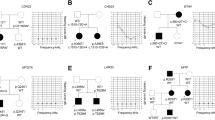
We recently described a novel missense variant [c.2090T>G:p.(Leu697Trp)] in the MYO3A gene, found in two Brazilian families with late-onset autosomal dominant nonsyndromic hearing loss (ADNSHL). Since then, with the objective of evaluating its contribution to ADNSHL in Brazil, the variant was screened in additional 101 pedigrees with probable ADNSHL without conclusive molecular diagnosis. The variant was found in three additional families, explaining 3/101 (~3%) of cases with ADNSHL in our Brazilian pedigree collection. In order to identify the origin of the variant, 21 individuals from the five families were genotyped with a high-density SNP array (~600 K SNPs— Axiom Human Origins; ThermoFisher). The identity by descent (IBD) approach revealed that many pairs of individuals from the different families have a kinship coefficient equivalent to that of second cousins, and all share a minimum haplotype of ~607 kb which includes the c.2090T>G variant suggesting it probably arose in a common ancestor. We inferred that the mutation occurred in a chromosomal segment of European ancestry and the time since the most common ancestor was estimated in 1100 years (CI = 775–1425). This variant was also reported in a Dutch family, which shares a 87,121 bp haplotype with the Brazilian samples, suggesting that Dutch colonists may have brought it to Northeastern Brazil in the 17th century. Therefore, the present study opens new avenues to investigate this variant not only in Brazilians but also in European families with ADNSHL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Morton CC, Nance WE. Newborn hearing screening—a silent revolution. N Engl J Med. 2006;354:2151–64.
Shearer AE, Hildebrand MS, Smith RJH. Hereditary hearing loss and deafness overview. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews. Seattle (WA): University of Washington; 1993-2019.
Smith RJ, Bale JF Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365:879–90.
Denoyelle F, Marlin S, Weil D, Moatti L, Chauvin P, Garabédian EN, et al. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: implications for genetic counselling. Lancet. 1999;353:1298–303.
Friedman TB, Griffith AJ. Human nonsyndromic sensorineural deafness. Annu Rev Genom Hum Genet. 2003;4:341–402.
Raviv D, Dror AA, Avraham KB. Hearing loss: a common disorder caused by many rare alleles. Ann N Y Acad Sci. 2010;1214:168–79.
Kalhammer G, Bähler M. Unconventional myosins. Essays Biochem. 2000;35:33–42.
Montell C, Rubin GM. The Drosophila ninaC locus encodes two photoreceptor cell specific proteins with domains homologous to protein kinases and the myosin heavy chain head. Cell. 1988;52:757–72.
Schneider ME, Dosé AC, Salles FT, Chang W, Erickson FL, Burnside B, et al. A new compartment at stereocilia tips defined by spatial and temporal patterns of myosin IIIa expression. J Neurosci. 2006;26:10243–52.
Raval MH, Quintero OA, Weck ML, Unrath WC, Gallagher JW, Cui R, et al. Impact of the motor and tail domains of class III Myosins on regulating the formation and elongation of actin protrusions. J Biol Chem. 2016;291:22781–92.
Salles FT, Merritt RC Jr, Manor U, Dougherty GW, Sousa AD, Moore JE, et al. Myosin IIIa boosts elongation of stereocilia by transporting espin 1 to the plus ends of actin filaments. Nat Cell Biol. 2009;11:443–50.
Walsh VL, Raviv D, Dror AA, Shahin H, Walsh T, Kanaan MN, et al. A mouse model for human hearing loss DFNB30 due to loss of function of myosin IIIA. Mamm Genome. 2011;22:170–7.
Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, et al. From flies’ eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA. 2002;99:7518–23.
Dosé AC, Burnside B. Cloning and chromosomal localization of a human class III myosin. Genomics 2000;67(Aug):333–42.
Grati M, Yan D, Raval MH, Walsh T, Ma Q, Chakchouk I, et al. MYO3A causes human dominant deafness and interacts with protocadherin 15-CD2 isoform. Hum Mutat. 2016;37:481–7.
Dantas VGL, Raval MH, Ballesteros A, Cui R, Gunther LK, Yamamoto GL, et al. Characterization of a novel MYO3A missense mutation associated with a dominant form of late onset hearing loss. Sci Rep. 2018;8:8706.
Salzano FM, Sans M. Interethnic admixture and the evolution of Latin American populations. Genet Mol Biol. 2014;37(1 Suppl):151–70.
de Farias AA, Nunes K, Lemes RB, Moura R, Fernandes GR, Melo US, et al. Origin and age of the causative mutations in KLC2, IMPA1, MED25 and WNT7A unravelled through Brazilian admixed populations. Sci Rep. 2018;8:16552.
Paskulin DD, Giacomazzi J, Achatz MI, Costa S, Reis RM, Hainaut P, et al. Ancestry of the Brazilian TP53 c.1010G>A (p.Arg337His, R337H) founder mutation: clues from haplotyping of short tandem repeats on chromosome 17p. PLoS ONE. 2015;10:e0143262.
Bampi GB, Bisso-Machado R, Hünemeier T, Gheno TC, Furtado GV, Veliz-Otani D, et al. Haplotype Study in SCA10 families provides further evidence for a common ancestral origin of the mutation. Neuromolecular Med. 2017;19:501–9.
Dias A, Lezirovitz K, Nicastro FS, Mendes B, Mingroni-Netto RC. Further evidence for loss-of-function mutations in the CEACAM16 gene causing nonsyndromic autosomal recessive hearing loss in humans. J Hum Genet. 2019;64:257–60.
Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, Painter I, et al. GWAS Tools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–31.
Nunes K, Zheng X, Torres M, Moraes ME, Piovezan BZ, Pontes GN, et al. HLA imputation in an admixed population: An assessment of the 1000 Genomes data as a training set. Hum Immunol. 2016;77:307–12.
Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012;28:3326–8.
Thornton T, Tang H, Hoffman TJ, Ochs-Balcom HM, Baan BJ, Risch N. Estimating kinship in admixed populations. Am J Hum Genet. 2012;91:122–38.
Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–81.
Gandolfo LC, Bahlo M, Speed TP. Dating rare mutations from small samples with dense marker data. Genetics. 2014;197:1315–27.
Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93:278–88.
Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Mata-Machado B. História do Sertão Noroeste de Minas Gerais (1690-930). Belo Horizonte: Imprensa Oficial; 1991.
Fundação João Pinheiro. História do café das matas de minas (1808-2015). Belo Horizonte: Fundação João Pinheiro; 2018. http://fjp.mg.gov.br/index.php/docman/dectec-2018/872-cafe-portal-com-capa/file.
Mello EC. O negócio do Brasil. São Paulo: Companhia das Letras; 2011.
Acknowledgements
The authors are indebted to Humberto C. Marcolino and to Maria Teresa B. M. Auricchio, for technical assistance. We thank Dr. Alfredo Tabith Jr. and all DERDIC staff for clinical assistance and Dr. Jeanne Oiticica for clinical supervision. We deeply thank Dr. Hans K. P. van Amstel, Dr. Rolph Pfundt, for the valuable information about the Dutch proband, Anna Morgan and Dr. Giorgia Girotto for information about Italian pedigrees and Dr. Miguel Angel Moreno Pelayo, about Spanish pedigrees with ADNHL. We are also grateful to FAPESP, CAPES, CNPq and NIH for their financial support. We thank all family members for their participation in the study.
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP - CEPID Human Genome and Stem Cell Research Center 2013/08028-1) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES. KN and DM were supported by United States National Institute of Health (NIH, R01 GM-075091).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bueno, A.S., Nunes, K., Dias, A.M.M. et al. Frequency and origin of the c.2090T>G p.(Leu697Trp) MYO3A variant associated with autosomal dominant hearing loss. Eur J Hum Genet 30, 13–21 (2022). https://doi.org/10.1038/s41431-021-00891-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-021-00891-0
This article is cited by
-
New year, new issue
European Journal of Human Genetics (2022)
-
Deafness—family matters
European Journal of Human Genetics (2022)
-
Genetic etiology of non-syndromic hearing loss in Latin America
Human Genetics (2022)
-
Molecular and genetic characterization of a large Brazilian cohort presenting hearing loss
Human Genetics (2022)