Abstract
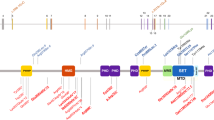
White–Sutton syndrome (WHSUS) is a neurodevelopmental disorder caused by heterozygous loss-of-function variants in POGZ. Through the Deciphering Developmental Disorders study and clinical testing, we identified 12 individuals from 10 families with pathogenic or likely pathogenic variants in POGZ (eight de novo and two inherited). Most individuals had delayed development and/or intellectual disability. We analyzed the clinical findings in our series and combined it with data from 89 previously reported individuals. The results demonstrate WHSUS is associated with variable developmental delay or intellectual disability, increased risk of obesity, visual defects, craniofacial dysmorphism, sensorineural hearing loss, feeding problems, seizures, and structural brain malformations. Our series includes further individuals with rod-cone dystrophy, cleft lip and palate, congenital diaphragmatic hernia, and duplicated renal drainage system, suggesting these are rare complications of WHSUS. In addition, we describe an individual with a novel, de novo missense variant in POGZ and features of WHSUS. Our work further delineates the phenotypic spectrum of WHSUS highlighting the variable severity of this disorder and the observation of familial pathogenic POGZ variants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Stessman HAF, Willemsen MH, Fenckova M, Penn O, Hoischen A, Xiong B, et al. Disruption of POGZ is associated with intellectual disability and autism spectrum disorders. Am J Hum Genet. 2016;98:541–52.
White J, Beck CR, Harel T, Posey JE, Jhangiani SN, Tang S, et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3.
Dentici ML, Niceta M, Pantaleoni F, Barresi S, Bencivenga P, Dallapiccola B, et al. Expanding the phenotypic spectrum of truncating POGZ mutations: association with CNS malformations, skeletal abnormalities, and distinctive facial dysmorphism. Am J Med Genet A 2017;173:1965–9.
Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BWM, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014;511:344–7.
Lelieveld SH, Reijnders MRF, Pfundt R, Yntema HG, Kamsteeg E-J, de Vries P, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6.
Ferretti A, Barresi S, Trivisano M, Ciolfi A, Dentici ML, Radio FC, et al. POGZ-related epilepsy: case report and review of the literature. Am J Med Genet A 2019;179:1631–6.
Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8.
Samanta D, Ramakrishnaiah R, Schaefer B. The neurological aspects related to POGZ mutation: case report and review of CNS malformations and epilepsy. Acta Neurol Belg. 2019;120:447–50.
Pascolini G, Agolini E, Fleischer N, Gulotta E, Cesario C, D’Elia G, et al. A novel patient with White-Sutton syndrome refines the mutational and clinical repertoire of the POGZ-related phenotype and suggests further observations. Am J Med Genet A. 2020;182:1791–5.
Assia Batzir N, Posey JE, Song X, Akdemir ZC, Rosenfeld JA, Brown CW, et al. Phenotypic expansion of POGZ-related intellectual disability syndrome (White-Sutton syndrome). Am J Med Genet A. 2020;182:38–52.
Garde A, Cornaton J, Sorlin A, Moutton S, Nicolas C, Juif C, et al. Neuropsychological study in 19 French patients with White-Sutton syndrome and POGZ mutations. Clin Genet. 2021;99:407–17.
Dal S, Hopper B, du Chattel MVR, Goel H A case of White-Sutton syndrome with previously described loss-of-function variant in DDE domain of POGZ (p.Arg1211*) and Kartagener syndrome. Am J Med Genet A. 2020;185:1006–7.
Nozawa R-S, Nagao K, Masuda H-T, Iwasaki O, Hirota T, Nozaki N, et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat Cell Biol. 2010;12:719–27.
Matsumura K, Seiriki K, Okada S, Nagase M, Ayabe S, Yamada I, et al. Pathogenic POGZ mutation causes impaired cortical development and reversible autism-like phenotypes. Nat Commun. 2020;11:859.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21.
Fukai R, Hiraki Y, Yofune H, Tsurusaki Y, Nakashima M, Saitsu H, et al. A case of autism spectrum disorder arising from a de novo missense mutation in POGZ. J Hum Genet. 2015;60:277–9.
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99.
Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84.
De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional, and chromatin genes disrupted in autism. Nature. 2014;515:209–15.
Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015;519:223–8.
Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet Lond Engl. 2015;385:1305–14.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med J Am Coll Med Genet. 2015;17:405–24.
Ellard S Best Practice Guidelines. The Association for Clinical Genomic Science. 2020. https://www.acgs.uk.com/quality/best-practice-guidelines/
Longoni M, High FA, Qi H, Joy MP, Hila R, Coletti CM, et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex Congenital Diaphragmatic Hernia. Hum Genet. 2017;136:679–91.
Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 2003;299:721–5.
Motamedi MR, Hong E-JE, Li X, Gerber S, Denison C, Gygi S, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–90.
Isidor B, Küry S, Rosenfeld JA, Besnard T, Schmitt S, Joss S, et al. De Novo Truncating Mutations in the Kinetochore-Microtubules Attachment Gene CHAMP1 Cause Syndromic Intellectual Disability. Hum Mutat. 2016;37:354–8.
Hempel M, Cremer K, Ockeloen CW, Lichtenbelt KD, Herkert JC, Denecke J, et al. De Novo Mutations in CHAMP1 Cause Intellectual Disability with Severe Speech Impairment. Am J Hum Genet. 2015;97:493–500.
Acknowledgements
We would like to thank the patients, families and clinicians who have contributed to this report. AEF was supported by the Wales Gene Park. Wales Gene Park is a Health and Care Research Wales funded infrastructure support group. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1 009-003]. This study makes use of DECIPHER (http://decipher.sanger.ac.uk), which is funded by Wellcome. See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgement. Clinical testing and analysis were performed by The Department of Medical Genetics, Telemark Hospital Trust, Skien, Norway and the Exeter Genomics Laboratory, Royal Devon & Exeter NHS Foundation Trust, UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Individuals who had evaluation or analysis beyond routine clinical care were part of research studies approved by the Cambridge South Research Ethics Committee (10/H0305/83).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Murch, O., Jain, V., Benneche, A. et al. Further delineation of the clinical spectrum of White–Sutton syndrome: 12 new individuals and a review of the literature. Eur J Hum Genet 30, 95–100 (2022). https://doi.org/10.1038/s41431-021-00961-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-021-00961-3
This article is cited by
-
White-Sutton syndrome and congenital heart disease: case report and literature review
BMC Pediatrics (2023)
-
Spectrum of congenital anomalies of the kidney and urinary tract (CAKUT) including renal parenchymal malformations during fetal life and the implementation of prenatal exome sequencing (WES)
Archives of Gynecology and Obstetrics (2023)
-
New year, new issue
European Journal of Human Genetics (2022)