Abstract
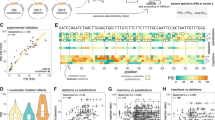
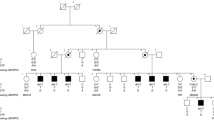
Pathogenic large inversions are rarely reported on DMD gene due to the lack of effective detection methods. Here we report two DMD pedigrees and proposed a reliable pipeline to define large inversions in DMD patients. In the first pedigree, conventional approaches including multiplex ligation-dependent probe amplification, and whole-exome sequencing by next generation sequencing were failed to detect any pathologic variant. Then an advanced analysis pipeline which consists of RNA-seq, cDNA array capture sequencing, optical mapping, long-read sequencing was built. RNA-seq and cDNA capture sequencing showed a complete absence of transcripts of exons 3–55. Optical mapping identified a 55 Mb pericentric inversion between Xp21 and Xq21. Subsequently, long-read sequencing and Sanger sequencing determined the inversion breakpoints at 32,915,769 and 87,989,324 of the X chromosomes. In the second pedigree, long-read sequencing was directly conducted and Sanger sequencing was performed to verify the mutation. Long-read sequencing and Sanger sequencing found breakpoints at 32,581,576 and 127,797,236 on DMD gene directly. In conclusion, large inversion might be a rare but important mutation type in DMD gene. An effective pipeline was built in detecting large inversion mutations based on long-read sequencing platforms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data are available from the corresponding author on reasonable request.
References
Burghes AH, Logan C, Hu X, Belfall B, Worton RG, Ray PN. A cDNA clone from the Duchenne/Becker muscular dystrophy gene. Nature. 1987;328:434–7.
Kunkel LM, Monaco AP, Middlesworth W, Ochs HD, Latt SA. Specific cloning of DNA fragments absent from the DNA of a male patient with an X chromosome deletion. Proc Natl Acad Sci USA. 1985;82:4778–82.
Emery AE. The muscular dystrophies. Lancet 2002;359:687–95.
Aartsma‐Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJB, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases that confirm the reading‐frame rule. Muscle Nerve: Off J Am Assoc Electrodiagn Med. 2006;34:135–44.
Rybakova IN, Patel JR, Ervasti JM. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J Cell Biol. 2000;150:1209–14.
Koenig M, Beggs A, Moyer M, Scherpf S, Heindrich K, Bettecken T, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498.
Tuffery‐Giraud S, Béroud C, Leturcq F, Yaou RB, Hamroun D, Michel‐Calemard L, et al. Genotype–phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD–DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30:934–45.
Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, et al. The TREAT‐NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36:395–402.
Tong YR, Geng C, Guan YZ, Zhao YH, Ren HT, Yao FX, et al. A Comprehensive Analysis of 2013 Dystrophinopathies in China: A Report From National Rare Disease Center. Front Neurol. 2020;11:572006.
Trippe H, Wieczorek S, Kötting J, Kress W, Schara U. Xp21/A translocation: a rarely considered genetic cause for manifesting carriers of duchenne muscular dystrophy. Neuropediatrics 2014;45:333–5.
Ligon AH, Kashork CD, Richards CS, Shaffer LG. Identification of female carriers for Duchenne and Becker muscular dystrophies using a FISH-based approach. Eur J Hum Genet. 2000;8:293–8.
Ishmukhametova A, Van Kien PK, Méchin D, Thorel D, Vincent M-C, Rivier F, et al. Comprehensive oligonucleotide array-comparative genomic hybridization analysis: new insights into the molecular pathology of the DMD gene. Eur J Hum Genet. 2012;20:1096–100.
van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The Third Revolution in Sequencing Technology. Trends Genet. 2018;34:666–81.
Zhang Q, Xu X, Ding L, Li H, Xu C, Gong Y, et al. Clinical application of single‐molecule optical mapping to a multigeneration FSHD1 pedigree. Mol Genet Genom Med. 2019;7:e565.
Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biol. 2013;14:405.
Loman NJ, Quick J, Simpson JT. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods. 2015;12:733–5.
Barseghyan H, Tang W, Wang RT, Almalvez M, Segura E, Bramble MS, et al. Next-generation mapping: a novel approach for detection of pathogenic structural variants with a potential utility in clinical diagnosis. Genome Med. 2017;9:90.
Mantere T, Kersten S, Hoischen A. Long-read sequencing emerging in medical genetics. Front Genet. 2019;10:426.
Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 2015;517:608–11.
Sanchis-Juan A, Stephens J, French CE, Gleadall N, Mégy K, Penkett C, et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short-and long-read genome sequencing. Genome Med. 2018;10:95.
Collins RL, Brand H, Redin CE, Hanscom C, Antolik C, Stone MR, et al. Defining the diverse spectrum of inversions, complex structural variation, and chromothripsis in the morbid human genome. Genome Biol. 2017;18:36.
Gonorazky H, Liang M, Cummings B, Lek M, Micallef J, Hawkins C, et al. RNA seq analysis for the diagnosis of muscular dystrophy. Ann Clin Transl Neurol. 2016;3:55–60.
Dai Y, Li P, Wang Z, Liang F, Yang F, Fang L, et al. Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet. 2020;57:109–20.
Deng J, Yu J, Li P, Luan X, Cao L, Zhao J, et al. Expansion of GGC Repeat in GIPC1 Is Associated with Oculopharyngodistal Myopathy. Am J Hum Genet. 2020;106:793–804.
Okubo M, Minami N, Goto K, Goto Y, Noguchi S, Mitsuhashi S, et al. Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: validation analysis of DMD mutations. J Hum Genet. 2016;61:483–9.
Lim BC, Lee S, Shin JY, Kim JI, Hwang H, Kim KJ, et al. Genetic diagnosis of Duchenne and Becker muscular dystrophy using next-generation sequencing technology: comprehensive mutational search in a single platform. J Med Genet. 2011;48:731–6.
Falzarano MS, Grilli A, Zia S, Fang M, Rossi R, Gualandi F, et al. RNA-seq in DMD urinary stem cells recognized muscle-related transcription signatures and addressed the identification of atypical mutations by whole-genome sequencing. HGG Adv. 2022;3:100054.
Weckselblatt B, Rudd MK. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 2015;31:587–99.
Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet. 2011;43:1074.
Fratter C, Dalgleish R, Allen SK, Santos R, Abbs S, Tuffery-Giraud S, et al. EMQN best practice guidelines for genetic testing in dystrophinopathies. Eur J Hum Genet. 2020;28:1141–59.
Acknowledgements
The authors express sincere gratitude to the patient and the family members who participated in the study. Special acknowledgements go to Dr Xiaoming (BGI Clinical Laboratory, Wuhan, China) for assistance for muscle tissue mRNA sequencing and to Ms Minming Zheng (Grandomics Biosciences, Beijing, China) for preparing the figures.
Funding
This study was funded by CAMS Innovation Fund for Medical Sciences (CIFMS) (No.2016-I2M-1-002) and Chinese National Basic Research Program (973) (No.2017YFC1001902).
Author information
Authors and Affiliations
Contributions
YD and JP designed the study. CG, CZ, YD, and JP collected clinical information of the pedigree. BZ and JH performed karyotype and PCR tests of family members. YZ and LC performed the pathology analysis of the muscle biopsy. PL, FL, and HJ conducted Bionano and ONT data analysis, YW and YRW performed Bionano and ONT sequencing and data collection, FY performed MLPA experiment. YD, JP LYC, DL, SL, DW, and LC supervised the project. CG, CZ, YT, MSP, YD, and JP drafted the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the ethics committee of the Peking Union Medical College Hospital (IRB #JS-1233). All participants and the parents of those <18years of age at the time of genetic testing gave written informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Geng, C., Zhang, C., Li, P. et al. Identification and characterization of two DMD pedigrees with large inversion mutations based on a long-read sequencing pipeline. Eur J Hum Genet 31, 504–511 (2023). https://doi.org/10.1038/s41431-022-01190-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01190-y
This article is cited by
-
Optical genome mapping identifies a balanced inversion disrupting DMD in a patient with Duchenne muscular dystrophy
Molecular Cytogenetics (2025)
-
Identifying inversions with breakpoints in the Dystrophin gene through long-read sequencing: report of two cases
BMC Medical Genomics (2024)
-
2023 in the European Journal of Human Genetics
European Journal of Human Genetics (2024)