Abstract
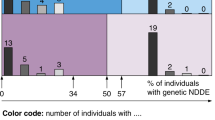
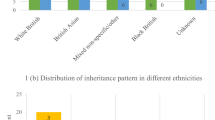
Pharmacoresistant epilepsy presenting during infancy poses both diagnostic and therapeutic challenges. We aim to identify diagnostic yield and treatment implications of exome sequencing (ES) as first-tier genetic testing for infantile-onset pharmacoresistant epilepsy. From June 2016 to December 2020, we enrolled patients with infantile-onset (age ≤ 12 months) pharmacoresistant epilepsy. 103 unrelated patients underwent ES. Clinical characteristics and changes in management due to the molecular diagnosis were studied. 42% (43/103) had epilepsy onset within the first month of life. After ES as first-tier genetic testing, 62% (64/103) of the cases were solved. Two partially solved cases (2%; 2/103) with heterozygous variants identified in ALDH7A1 known to cause autosomal recessive pyridoxine dependent epilepsy underwent genome sequencing (GS). Two novel large deletions in ALDH7A1 were detected in both cases. ES identified 66 pathogenic and likely pathogenic single nucleotide variants (SNVs) in 27 genes. 19 variants have not been previously reported. GS identified two additional copy number variations (CNVs). The most common disease-causing genes are SCN1A (13%; 13/103) and KCNQ2 (8%; 8/103). Eight percent (8/103) of the patients had treatable disorders and specific treatments were provided resulting in seizure freedom. Pyridoxine dependent epilepsy was the most common treatable epilepsy (6%; 6/103). Furthermore, 35% (36/103) had genetic defects which guided gene-specific treatments. Altogether, the diagnostic yield is 64%. Molecular diagnoses change management in 43% of the cases. This study substantiates the use of next generation sequencing (NGS) as the first-tier genetic investigation in infantile-onset pharmacoresistant epilepsy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Symonds JD, Zuberi SM, Stewart K, McLellan A, O’Regan M, MacLeod S, et al. Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain. 2019;142:2303–18.
Gaily E, Lommi M, Lapatto R, Lehesjoki AE. Incidence and outcome of epilepsy syndromes with onset in the first year of life: A retrospective population-based study. Epilepsia. 2016;57:1594–601.
Stödberg T, Tomson T, Barbaro M, Stranneheim H, Anderlid BM, Carlsson S, et al. Epilepsy syndromes, etiologies, and the use of next-generation sequencing in epilepsy presenting in the first 2 years of life: A population-based study. Epilepsia. 2020;61:2486–99.
Laxer KD, Trinka E, Hirsch LJ, Cendes F, Langfitt J, Delanty N, et al. The consequences of refractory epilepsy and its treatment. Epilepsy Behav. 2014;37:59–70.
Eltze CM, Chong WK, Cox T, Whitney A, Cortina-Borja M, Chin RF, et al. A population-based study of newly diagnosed epilepsy in infants. Epilepsia. 2013;54:437–45.
Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia. 2010;51:676–85.
Allen NM, Conroy J, Shahwan A, Ennis S, Lynch B, Lynch SA, et al. Chromosomal microarray in unexplained severe early onset epilepsy - A single centre cohort. Eur J Paediatr Neurol. 2015;19:390–4.
Butler KM, da Silva C, Alexander JJ, Hegde M, Escayg A. Diagnostic Yield From 339 Epilepsy Patients Screened on a Clinical Gene Panel. Pediatr Neurol. 2017;77:61–66.
Allen NM, Conroy J, Shahwan A, Lynch B, Correa RG, Pena SD, et al. Unexplained early onset epileptic encephalopathy: Exome screening and phenotype expansion. Epilepsia. 2016;57:e12–17.
Zhou P, He N, Zhang JW, Lin ZJ, Wang J, Yan LM, et al. Novel mutations and phenotypes of epilepsy-associated genes in epileptic encephalopathies. Genes Brain Behav. 2018;17:e12456.
Olson HE, Kelly M, LaCoursiere CM, Pinsky R, Tambunan D, Shain C, et al. Genetics and genotype-phenotype correlations in early onset epileptic encephalopathy with burst suppression. Ann Neurol. 2017;81:419–29.
Segal E, Pedro H, Valdez-Gonzalez K, Parisotto S, Gliksman F, Thompson S, et al. Diagnostic Yield of Epilepsy Panels in Children With Medication-Refractory Epilepsy. Pediatr Neurol. 2016;64:66–71.
Mercimek-Mahmutoglu S, Patel J, Cordeiro D, Hewson S, Callen D, Donner EJ, et al. Diagnostic yield of genetic testing in epileptic encephalopathy in childhood. Epilepsia. 2015;56:707–16.
Zhang Y, Kong W, Gao Y, Liu X, Gao K, Xie H, et al. Gene Mutation Analysis in 253 Chinese Children with Unexplained Epilepsy and Intellectual/Developmental Disabilities. PLoS One. 2015;10:e0141782.
Ostrander BEP, Butterfield RJ, Pedersen BS, Farrell AJ, Layer RM, Ward A, et al. Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy. NPJ Genom Med. 2018;3:22.
Demos M, Guella I, DeGuzman C, McKenzie MB, Buerki SE, Evans DM, et al. Diagnostic Yield and Treatment Impact of Targeted Exome Sequencing in Early-Onset Epilepsy. Front Neurol. 2019;10:434.
Lemke JR, Riesch E, Scheurenbrand T, Schubach M, Wilhelm C, Steiner I, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–98.
Trump N, McTague A, Brittain H, Papandreou A, Meyer E, Ngoh A, et al. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. J Med Genet. 2016;53:310–7.
Ortega-Moreno L, Giraldez BG, Soto-Insuga V, Losada-Del Pozo R, Rodrigo-Moreno M, Alarcon-Morcillo C, et al. Molecular diagnosis of patients with epilepsy and developmental delay using a customized panel of epilepsy genes. PLoS One. 2017;12:e0188978.
Carvill GL, Heavin SB, Yendle SC, McMahon JM, O’Roak BJ, Cook J, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825–30.
Michaud JL, Lachance M, Hamdan FF, Carmant L, Lortie A, Diadori P, et al. The genetic landscape of infantile spasms. Hum Mol Genet. 2014;23:4846–58.
Costain G, Cordeiro D, Matviychuk D, Mercimek-Andrews S. Clinical Application of Targeted Next-Generation Sequencing Panels and Whole Exome Sequencing in Childhood Epilepsy. Neuroscience. 2019;418:291–310.
Ko A, Youn SE, Kim SH, Lee JS, Kim S, Choi JR, et al. Targeted gene panel and genotype-phenotype correlation in children with developmental and epileptic encephalopathy. Epilepsy Res. 2018;141:48–55.
Kothur K, Holman K, Farnsworth E, Ho G, Lorentzos M, Troedson C, et al. Diagnostic yield of targeted massively parallel sequencing in children with epileptic encephalopathy. Seizure. 2018;59:132–40.
Snoeijen-Schouwenaars FM, van Ool JS, Verhoeven JS, van Mierlo P, Braakman HMH, Smeets EE, et al. Diagnostic exome sequencing in 100 consecutive patients with both epilepsy and intellectual disability. Epilepsia. 2019;60:155–64.
Hoelz H, Herdl C, Gerstl L, Tacke M, Vill K, von Stuelpnagel C, et al. Impact on Clinical Decision Making of Next-Generation Sequencing in Pediatric Epilepsy in a Tertiary Epilepsy Referral Center. Clin EEG Neurosci. 2020;51:61–69.
Johannessen Landmark C, Potschka H, Auvin S, Wilmshurst JM, Johannessen SI, Kasteleijn-Nolst Trenite D, et al. The role of new medical treatments for the management of developmental and epileptic encephalopathies: Novel concepts and results. Epilepsia. 2021.
Devinsky O, Vezzani A, O’Brien TJ, Jette N, Scheffer IE, de Curtis M, et al. Epilepsy. Nat Rev Dis Prim. 2018;4:18024.
Almannai M, El-Hattab AW. Inborn Errors of Metabolism with Seizures: Defects of Glycine and Serine Metabolism and Cofactor-Related Disorders. Pediatr Clin North Am. 2018;65:279–99.
Sharma S, Prasad AN. Inborn Errors of Metabolism and Epilepsy: Current Understanding, Diagnosis, and Treatment Approaches. Int J Mol Sci. 2017;18:1384.
Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2010;51:1069–77.
Kosugi S, Momozawa Y, Liu X, Terao C, Kubo M, Kamatani Y. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing. Genome Biol. 2019;20:117.
Zhu M, Need AC, Han Y, Ge D, Maia JM, Zhu Q, et al. Using ERDS to infer copy-number variants in high-coverage genomes. Am J Hum Genet. 2012;91:408–21.
Shotelersuk V, Wichadakul D, Ngamphiw C, Srichomthong C, Phokaew C, Wilantho A, et al. The Thai reference exome (T-REx) variant database. Clin Genet. 2021;100:703–12.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020;22:245–57.
Arafat A, Jing P, Ma Y, Pu M, Nan G, Fang H, et al. Unexplained Early Infantile Epileptic Encephalopathy in Han Chinese Children: Next-Generation Sequencing and Phenotype Enriching. Sci Rep. 2017;7:46227.
Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am J Hum Genet. 2017;101:664–85.
Tan TY, Lunke S, Chong B, Phelan D, Fanjul-Fernandez M, Marum JE, et al. A head-to-head evaluation of the diagnostic efficacy and costs of trio versus singleton exome sequencing analysis. Eur J Hum Genet. 2019;27:1791–9.
Gao C, Wang X, Mei S, Li D, Duan J, Zhang P, et al. Diagnostic Yields of Trio-WES Accompanied by CNVseq for Rare Neurodevelopmental Disorders. Front Genet. 2019;10:485.
Li J, Gao K, Yan H, Xiangwei W, Liu N, Wang T, et al. Reanalysis of whole exome sequencing data in patients with epilepsy and intellectual disability/mental retardation. Gene. 2019;700:168–75.
Rochtus A, Olson HE, Smith L, Keith LG, El Achkar C, Taylor A, et al. Genetic diagnoses in epilepsy: The impact of dynamic exome analysis in a pediatric cohort. Epilepsia. 2020;61:249–58.
Bok LA, Maurits NM, Willemsen MA, Jakobs C, Teune LK, Poll-The BT, et al. The EEG response to pyridoxine-IV neither identifies nor excludes pyridoxine-dependent epilepsy. Epilepsia. 2010;51:2406–11.
Kanno J, Kure S, Narisawa A, Kamada F, Takayanagi M, Yamamoto K, et al. Allelic and non-allelic heterogeneities in pyridoxine dependent seizures revealed by ALDH7A1 mutational analysis. Mol Genet Metab. 2007;91:384–9.
Plecko B, Paul K, Paschke E, Stoeckler-Ipsiroglu S, Struys E, Jakobs C, et al. Biochemical and molecular characterization of 18 patients with pyridoxine-dependent epilepsy and mutations of the antiquitin (ALDH7A1) gene. Hum Mutat. 2007;28:19–26.
Mefford HC, Zemel M, Geraghty E, Cook J, Clayton PT, Paul K, et al. Intragenic deletions of ALDH7A1 in pyridoxine-dependent epilepsy caused by Alu-Alu recombination. Neurology. 2015;85:756–62.
Perez B, Gutierrez-Solana LG, Verdu A, Merinero B, Yuste-Checa P, Ruiz-Sala P, et al. Clinical, biochemical, and molecular studies in pyridoxine-dependent epilepsy. Antisense therapy as possible new therapeutic option. Epilepsia. 2013;54:239–48.
Clark MM, Hildreth A, Batalov S, Ding Y, Chowdhury S, Watkins K, et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci Transl Med 2019;11:eaat6177.
Farnaes L, Hildreth A, Sweeney NM, Clark MM, Chowdhury S, Nahas S, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med. 2018;3:10.
Kamolvisit W, Phowthongkum P, Boonsimma P, Kuptanon C, Rojnueangnit K, Wattanasirichaigoon D, et al. Rapid exome sequencing as the first-tier investigation for diagnosis of acutely and severely ill children and adults in Thailand. Clin Genet. 2021;100:100–5.
Acknowledgements
We would like to thank the patients and their families for participating in this study. We also thank Natrujee Wiwattanadittakul M.D., Chinnuwat Sanguansermsri M.D., Kamornwan Katanyuwong M.D., Division of Neurology, Department of Pediatrics, Maharaj Nakorn Chiang Mai Hospital; Somjit Sri-Udomkajorn M.D., Thanin Wechapinan M.D., Sirorat Suwannachote M.D., Chulaluck Kuptanon M.D., Department of Pediatrics, Queen Sirikit National Institute of Child Health, Surachai Likasitwattankul, M.D., Sorawit Viravan, M.D., Mongkon Chanvanichtrakool, M.D. Department of Pediatrics, Siriraj Hospital; Thitiporn Fangsaad, M.D. Department of Pediatrics, Bhumibol Adulyadej Hospital; Vitchayaporn Emarach Saengow, M.D. Department of Pediatrics, Maharat Nakhon Ratchasima Hospital, for their excellent care of the patients.
Funding
The study was supported by Health Systems Research Institute (65-040, 64-125).
Author information
Authors and Affiliations
Contributions
PB, WK, SP and TD collected data. WC, CP, CI, and RI performed DNA sequencing and analysis. PB and CI analyzed the data, drafted and revised the paper. KS and VS revised the draft. KS and VS conceived the study and obtained funding. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University, Thailand (IRB No. 264/62). Written informed consent was obtained from parents or legal guardians of the participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boonsimma, P., Ittiwut, C., Kamolvisit, W. et al. Exome sequencing as first-tier genetic testing in infantile-onset pharmacoresistant epilepsy: diagnostic yield and treatment impact. Eur J Hum Genet 31, 179–187 (2023). https://doi.org/10.1038/s41431-022-01202-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01202-x
This article is cited by
-
Advancing precision public health at Genomics Thailand
Nature Health (2026)
-
Genetic etiology of 283 Chinese individuals with epilepsy using copy number variation sequencing and whole exome sequencing: a single-center cohort study
BMC Medical Genomics (2025)
-
Propofol rescues voltage-dependent gating of HCN1 channel epilepsy mutants
Nature (2024)
-
The value of exomes across the ages
European Journal of Human Genetics (2023)
-
Nine patients with KCNQ2-related neonatal seizures and functional studies of two missense variants
Scientific Reports (2023)