Abstract
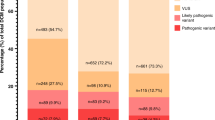
It was previously suggested that increasing the number of genes on diagnostic gene panels could increase the genetic yield in patient with dilated cardiomyopathy (DCM). We explored the diagnostic and prognostic relevance of testing DCM patients with an expanded gene panel. The current study included 225 consecutive DCM patients who had no genetic diagnosis after a 48-gene cardiomyopathy-panel. These were then evaluated using an expanded gene panel of 299 cardiac-associated genes. A likely pathogenic/pathogenic (P/LP) variant was detected in 13 patients. Five variants were reclassifications of variants found in genes which were already detected using the 48 gene panel. Only one of the other eight variants could explain the phenotype of the patient (KCNJ2). The panel detected 186 VUSs in 127 patients (of which 6 also had a P/LP variant). The presence of a VUS was significantly associated with the combined end-point of mortality, heart failure hospitalization, heart transplantation or life-threatening arrhythmias(HR, 2.04 [95% CI, 1.15 to 3.65]; p = 0.02). The association of a VUS with prognosis remained when we only included VUSs in robust DCM-associated genes (high suspicious VUSs), but disappeared when we only included VUSs in non-robust DCM-associated genes (low suspicious VUSs), highlighting the importance of weighing of VUSs. Overall, the use of large gene panels for genetic testing in DCM does not increase the diagnostic yield, although a VUS in a robust DCM-associated gene is associated with an adverse prognosis. Altogether, current diagnostic gene panels should be limited to the robust DCM-associated genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Bohm M, et al. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J. 2016;37:1850–8.
Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, et al. Genetic evaluation of cardiomyopathy-A heart failure society of America practice guideline. J Card Fail. 2018;24:281–302.
Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, et al. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American college of medical genetics and genomics (ACMG). Genet Med: Off J Am Coll Med Genet. 2018;20:899–909.
McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Böhm M, et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2021;42:3599–726.
Verdonschot JAJ, Hazebroek MR, Krapels IPC, Henkens M, Raafs A, Wang P, et al. Implications of Genetic Testing in Dilated Cardiomyopathy. Circ Genom Precis Med. 2020;13:476–87.
Jordan E, Peterson L, Ai T, Asatryan B, Bronicki L, Brown E, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation. 2021;144:7–19.
Mazzarotto F, Tayal U, Buchan RJ, Midwinter W, Wilk A, Whiffin N, et al. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation. 2020;141:387–98.
Stroeks S, Hellebrekers D, Claes GRF, Tayal U, Krapels IPC, Vanhoutte EK, et al. Clinical impact of re-evaluating genes and variants implicated in dilated cardiomyopathy. Genetics in medicine: Off J Am Coll Med Genet. 2021;23:2186–93.
Harakalova M, Kummeling G, Sammani A, Linschoten M, Baas AF, van der Smagt J, et al. A systematic analysis of genetic dilated cardiomyopathy reveals numerous ubiquitously expressed and muscle-specific genes. Eur J heart Fail. 2015;17:484–93.
Walsh R, Offerhaus JA, Tadros R, Bezzina CR. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat Rev Cardiol. 2022;19:151–67.
Walsh R, Tadros R, Bezzina CR. When genetic burden reaches threshold. Eur Heart J. 2020;41:3849–55.
Henkens M, Weerts J, Verdonschot JAJ, Raafs AG, Stroeks S, Sikking MA, et al. Improving diagnosis and risk stratification across the ejection fraction spectrum: the Maastricht Cardiomyopathy registry. ESC Heart Fail. 2022;9:1463–70.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med: Off J Am Coll Med Genet. 2015;17:405–24.
Morales A, Kinnamon DD, Jordan E, Platt J, Vatta M, Dorschner MO, et al. Variant interpretation for dilated cardiomyopathy: refinement of the american college of medical genetics and genomics/clingen guidelines for the DCM precision medicine study. Circ Genom Precis Med. 2020;13:e002480.
Porto G, Brissot P, Swinkels DW, Zoller H, Kamarainen O, Patton S, et al. EMQN best practice guidelines for the molecular genetic diagnosis of hereditary hemochromatosis (HH). Eur J Hum Genet: EJHG. 2016;24:479–95.
Zeppenfeld K, Tfelt-Hansen J, de Riva M, Winkel BG, Behr ER, Blom NA, et al. 2022 ESC guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur Heart J. 2022;43:3997–126.
Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med: Off J Am Coll Med Genet. 2015;17:880–8.
Mazzarotto F, Girolami F, Boschi B, Barlocco F, Tomberli A, Baldini K, et al. Defining the diagnostic effectiveness of genes for inclusion in panels: the experience of two decades of genetic testing for hypertrophic cardiomyopathy at a single center. Genet Med: Off J Am Coll Med Genet. 2019;21:284–92.
Schobers G, Schieving JH, Yntema HG, Pennings M, Pfundt R, Derks R, et al. Reanalysis of exome negative patients with rare disease: a pragmatic workflow for diagnostic applications. Genome Med. 2022;14:66.
Bourfiss M, van Vugt M, Alasiri AI, Ruijsink B, van Setten J, Schmidt AF, et al. Penetrance and disease expression of (likely) pathogenic variants associated with inherited cardiomyopathies in the general population. Circ Genom Precis Med. 2022;15:e003704.
Lazarte J, Jurgens SJ, Choi SH, Khurshid S, Morrill VN, Weng LC, et al. LMNA variants and risk of adult-onset cardiac disease. J Am Coll Cardiol. 2022;80:50–9.
Pirruccello JP, Bick A, Wang M, Chaffin M, Friedman S, Yao J, et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat Commun. 2020;11:2254.
Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, et al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the sarcomeric human cardiomyopathy registry (SHaRe). Circulation. 2018;138:1387–98.
Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, et al. Reappraisal of reported genes for sudden arrhythmic death: evidence-based evaluation of gene validity for brugada syndrome. Circulation. 2018;138:1195–205.
Tadros R, Francis C, Xu X, Vermeer AMC, Harper AR, Huurman R, et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat Genet. 2021;53:128–34.
Funding
JV is supported by a Dutch Heart Foundation Dekker – Clinical Scientist grant. The views expressed in this work are those of the authors and not necessarily those of the funders
Author information
Authors and Affiliations
Contributions
Conceptualization: SS, DH, JV; Data curation: SS, DH, GC, MH, MS, JV; Formal Analysis: SS, JV; Funding acquisition: JV, HB, A-vdW; Investigation: SS, DH, GC, IK, EV, AH-vdE, JV; Methodology: JV, Resources: HB, A-vdW; Supervision: HB, JV; Visualization: SS, JV; Writing – original draft: SS, DH, JV; Writing – review & editing: GC, IK, MH, MS, EV, AH-vdE.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was performed according to the declaration of Helsinki and was approved by the institutional Medical Ethics Committee of the Maastricht University Medical Center. All patients gave written informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Stroeks, S.L.V.M., Hellebrekers, D., Claes, G.R.F. et al. Diagnostic and prognostic relevance of using large gene panels in the genetic testing of patients with dilated cardiomyopathy. Eur J Hum Genet 31, 776–783 (2023). https://doi.org/10.1038/s41431-023-01384-y
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-023-01384-y
This article is cited by
-
Investigating the dual role of mitochondrial and nuclear genome variants in pediatric cardiomyopathies
Scientific Reports (2025)
-
High societal costs and reduced health-related quality of life in inflammatory and systemic immune disease-associated dilated cardiomyopathies
Quality of Life Research (2025)
-
Unusual genomic variants require unusual analyses
European Journal of Human Genetics (2023)